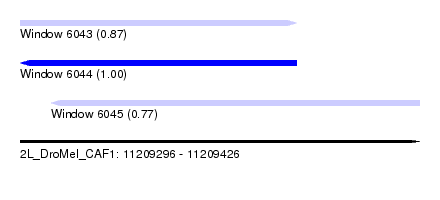
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,209,296 – 11,209,426 |
| Length | 130 |
| Max. P | 0.999870 |

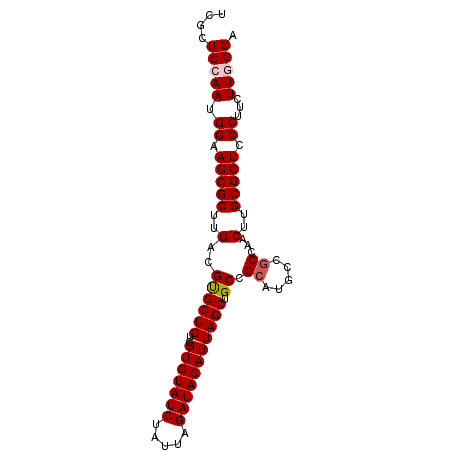
| Location | 11,209,296 – 11,209,386 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -24.38 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
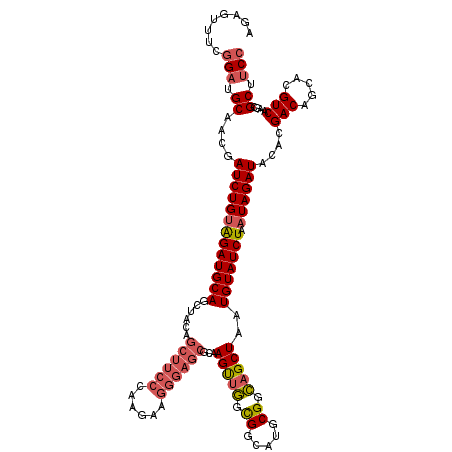
>2L_DroMel_CAF1 11209296 90 + 22407834 UCGCUCCAAUGGAAGCGCUUGACGUGCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCACCAACUUGCGCUCCCUUUUUGGGA ....(((((.((.(((((.((.(((((.((.(((((((.....))))))).....)).))))).)).......))))).))...))))). ( -27.70) >DroSec_CAF1 45166 90 + 1 UCGCUCCAAUGGAAGCGCUUGACGUGCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUCUUGGGA ....(((((.((.(((((..(....((((..(((((((.....))))))))))).((.(....).))...)..))))).))...))))). ( -27.50) >DroSim_CAF1 47056 90 + 1 UCGCUCCAAUGGAAGCGCUUGACGUGCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUCUUGGGA ....(((((.((.(((((..(....((((..(((((((.....))))))))))).((.(....).))...)..))))).))...))))). ( -27.50) >DroEre_CAF1 45692 89 + 1 UCGCUCCAAUGGAAGCGCUUGACGCGCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUU-UUUGGA ....(((((.((.(((((..(..((((((..(((((((.....))))))))))).)).((.....))...)..))))).))..-.))))) ( -30.00) >DroYak_CAF1 47200 90 + 1 UCGCUCCAAUGGAAGCGCUUGACGUGCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUCUUUGGA ....(((((.((.(((((..(....((((..(((((((.....))))))))))).((.(....).))...)..))))).))....))))) ( -27.70) >consensus UCGCUCCAAUGGAAGCGCUUGACGUGCUGUCGUGUAUCUAUUAGAUACAUUAGCUGCCGCAUGCCGCCAACUUGCGCUCCCUUCUUGGGA ....(((((.((.(((((..(..((((((..(((((((.....))))))))))).)).((.....))...)..))))).))...))))). (-24.38 = -24.82 + 0.44)

| Location | 11,209,296 – 11,209,386 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -32.74 |
| Energy contribution | -32.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.32 |
| SVM RNA-class probability | 0.999870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11209296 90 - 22407834 UCCCAAAAAGGGAGCGCAAGUUGGUGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGA ..((((...((((((((.(((((.((.....)).)))))..((((((.....)))))).(((.....)))..)))))))).))))..... ( -32.60) >DroSec_CAF1 45166 90 - 1 UCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGA ..((((...((((((((.(((((.((.....)).)))))..((((((.....)))))).(((.....)))..)))))))).))))..... ( -35.80) >DroSim_CAF1 47056 90 - 1 UCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGA ..((((...((((((((.(((((.((.....)).)))))..((((((.....)))))).(((.....)))..)))))))).))))..... ( -35.80) >DroEre_CAF1 45692 89 - 1 UCCAAA-AAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCGCGUCAAGCGCUUCCAUUGGAGCGA (((((.-..((((((((.(((((.((.....)).)))))..((((((.....)))))).(((.....)))..)))))))).))))).... ( -36.00) >DroYak_CAF1 47200 90 - 1 UCCAAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGA (((((....((((((((.(((((.((.....)).)))))..((((((.....)))))).(((.....)))..)))))))).))))).... ( -35.40) >consensus UCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCCAUUGGAGCGA (((.((...((((((((.(((((.((.....)).)))))..((((((.....)))))).(((.....)))..)))))))).))))).... (-32.74 = -32.58 + -0.16)

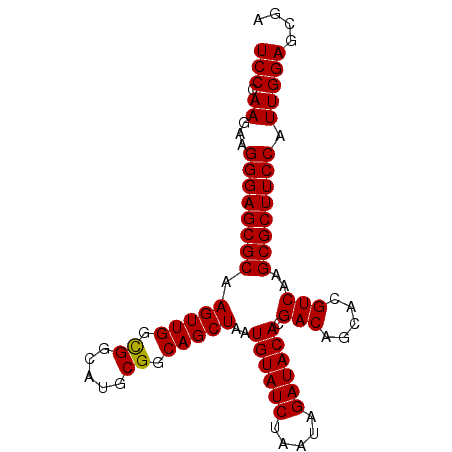

| Location | 11,209,306 – 11,209,426 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -33.01 |
| Energy contribution | -33.98 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11209306 120 - 22407834 AGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAAAAGGGAGCGCAAGUUGGUGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCC ........(((.((....((((((((((((((((((....((((.....))))((....))..)))))..((....))...))))))).))))))....(((.....)))....)).))) ( -37.80) >DroSec_CAF1 45176 120 - 1 AAAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCC ........(((.((....(((((((((((((......((((((......))))))...(((((.((.....)).)))))..))))))).))))))....(((.....)))....)).))) ( -38.70) >DroSim_CAF1 47066 120 - 1 AGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCC ........(((.((....(((((((((((((......((((((......))))))...(((((.((.....)).)))))..))))))).))))))....(((.....)))....)).))) ( -38.70) >DroEre_CAF1 45702 112 - 1 -------CGGAUGCAGCGAUCUGUGGAUGCAGCCGCAGCUUCCAAA-AAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCGCGUCAAGCGCUUCC -------.(((...((((..((...(((((.((((((((((((...-..))))))((......))....)))))).(((..((((((.....))))))....)))))))).))))))))) ( -45.30) >DroYak_CAF1 47210 120 - 1 AGAGUUUCGGUUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCAAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCC .(((((..(((((((.(........).)))))))..)))))........((((((((.(((((.((.....)).)))))..((((((.....)))))).(((.....)))..)))))))) ( -42.50) >DroAna_CAF1 41544 100 - 1 ---------GAUGCAACGAUCUGAAGAUGCAGUAUCU-----------GGGGAUCUCUAGCCAGCGGCAUGCGGUAGCUAAUGUAUCUAAUAGAUACACGACAGCAUGUCAAACGCUUCC ---------((.((...(((((..(((((...)))))-----------..)))))..........(((((((.((......((((((.....))))))..)).)))))))....)).)). ( -28.80) >consensus AGAGUUUCGGAUGCAACGAUCUGUAGAUGCAGCUACAGCUUCCCAAGAAGGGAGCGCAAGUUGGCGGCAUGCGGCAGCUAAUGUAUCUAAUAGAUACACGACAGCACGUCAAGCGCUUCC ........(((.((....(((((((((((((......((((((......))))))...(((((.((.....)).)))))..))))))).))))))....(((.....)))....)).))) (-33.01 = -33.98 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:37 2006