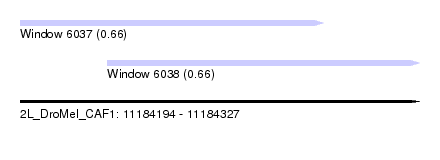
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,184,194 – 11,184,327 |
| Length | 133 |
| Max. P | 0.657716 |

| Location | 11,184,194 – 11,184,295 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -17.34 |
| Energy contribution | -18.43 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
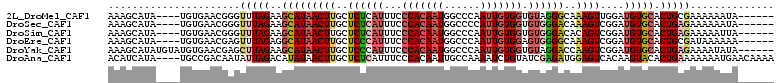
>2L_DroMel_CAF1 11184194 101 + 22407834 AAAGCAUA----UGUGAACGGGUUUAGAAGCAUAACUUGCUCUCAUUUCCCACAAUGGCCCAAUUGUGGUGUAGGGCAAAGUUGGAUGUGCACUGCGAAAAAAUA------ ...(((((----(......(((....((((((.....)))).))....))).((((.((((.((......)).))))...)))).))))))..............------ ( -27.20) >DroSec_CAF1 20695 101 + 1 AAAGCAUA----UGUGAACGGGUUUAGAAGCAUAACUUGCUCUCAUUUCCCACAAUGGCCCCAUUGUGGUGUGGGACAAAGUCGGAUGUGCACUGAGAAAAAAUA------ ...(((((----(.(((....((.....((((.....))))(((((...((((((((....)))))))).)))))))....))).))))))..............------ ( -31.60) >DroSim_CAF1 20469 101 + 1 AAAGCAUA----UGUGAACGGGUUUAGAAGCAUAACUUGCUCUCAUUUCCCACAAUGGCCCAAUUGUGGUGUGGGACACAGUCGGAUGUGCACUGAGAAAAAUUA------ ...(((((----(.(((...(........(((.....)))((((((...(((((((......))))))).))))))..)..))).))))))..............------ ( -28.50) >DroEre_CAF1 21318 101 + 1 AAAGCAUA----UGUGAACGAGUUUAGAGGCAUAACUUGCUCCCAUUUCCCACAAUGGCCCAAUUGUGGAGUGGGGCAAAGUCGGAUGUGCACUGCGAUAAAAAA------ ...(((((----(.(((....(((....))).....((((.((((((..(((((((......)))))))))))))))))..))).))))))..............------ ( -32.30) >DroYak_CAF1 22048 105 + 1 AAAGCAUAUGUAUGUGAACGAGCUUAGAAGCAUAACUUGCUCCCAUUUCCCACAAUGGCCCAAUUGUGGUGUAGGACCAAGUCGGAUGUGCACUGAGAAAAUAUA------ ...((((....)))).......(((((..((((((((((.(((.((...(((((((......))))))).)).))).)))))....))))).)))))........------ ( -27.10) >DroAna_CAF1 18506 107 + 1 ACAUCAUA----UGCCGACAAUAUUAGACAUAUAACUUGCUCUCAUUUCCCACAAUUGCCAAAAUCUGUAUCGAGAUGGAGUCACAAGUACACUGAAAAAAAUGAACAAAA ...(((((----((.(..........).))))).((((((((.((((((..((((((.....))).)))...)))))))))...))))).....))............... ( -13.80) >consensus AAAGCAUA____UGUGAACGAGUUUAGAAGCAUAACUUGCUCUCAUUUCCCACAAUGGCCCAAUUGUGGUGUGGGACAAAGUCGGAUGUGCACUGAGAAAAAAUA______ ......................(((((..(((((((((..((((((...(((((((......))))))).))))))..))))....))))).))))).............. (-17.34 = -18.43 + 1.09)

| Location | 11,184,223 – 11,184,327 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.30 |
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -12.91 |
| Energy contribution | -13.55 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11184223 104 + 22407834 AACUUGCUCUCAUUUCCCACAAUGGCCCAAUUGUGGUGUAGGGCAAAGUUGGAUGUGCACUGCGAAAAAAUA-------UUUUACAUAA-------AGUUUCCUAGUCAUGAUAACAA ...(((((((((....(((((((......))))))))).))))))).((((...(((.((((.((((...((-------(.....))).-------..)))).)))))))..)))).. ( -25.30) >DroSec_CAF1 20724 102 + 1 AACUUGCUCUCAUUUCCCACAAUGGCCCCAUUGUGGUGUGGGACAAAGUCGGAUGUGCACUGAGAAAAAAUA-------UUUGAAAGGA----U--AAUAUCAU---AAUGAUAACAA ...(((.((((((...((((((((....)))))))).)))))))))..((((((((.............)))-------))))).....----.--..((((..---...)))).... ( -25.52) >DroSim_CAF1 20498 88 + 1 AACUUGCUCUCAUUUCCCACAAUGGCCCAAUUGUGGUGUGGGACACAGUCGGAUGUGCACUGAGAAAAAUUA-------UUUGC-----------------------CAUGAUAACAA .......(((((..(((((((.(.((......)).))))))))((((......))))...)))))....(((-------((...-----------------------...)))))... ( -19.80) >DroEre_CAF1 21347 84 + 1 AACUUGCUCCCAUUUCCCACAAUGGCCCAAUUGUGGAGUGGGGCAAAGUCGGAUGUGCACUGCGAUAAAAAA-------CU---------------------------AUCAUAAAAA ...((((.((((((..(((((((......))))))))))))))))).((((.(.(....)).))))......-------..---------------------------.......... ( -23.70) >DroYak_CAF1 22081 104 + 1 AACUUGCUCCCAUUUCCCACAAUGGCCCAAUUGUGGUGUAGGACCAAGUCGGAUGUGCACUGAGAAAAUAUA-------UUUACCACGA-------AGUUUUUUGAUCUUCAUAGAAA ..((((.(((.((...(((((((......))))))).)).))).))))((((.......)))).........-------........((-------((.........))))....... ( -20.20) >DroAna_CAF1 18535 117 + 1 AACUUGCUCUCAUUUCCCACAAUUGCCAAAAUCUGUAUCGAGAUGGAGUCACAAGUACACUGAAAAAAAUGAACAAAACUCUAAUAUGAAGACUAAACUUUACU-GCCAGAAUUAUAA .((((((((.((((((..((((((.....))).)))...)))))))))...)))))...(((........((.......)).....(((((......)))))..-..)))........ ( -14.40) >consensus AACUUGCUCUCAUUUCCCACAAUGGCCCAAUUGUGGUGUGGGACAAAGUCGGAUGUGCACUGAGAAAAAAUA_______UUUAA_A_GA_______A_UUU__U___CAUGAUAACAA .((((..((((((...(((((((......))))))).))))))..))))..................................................................... (-12.91 = -13.55 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:30 2006