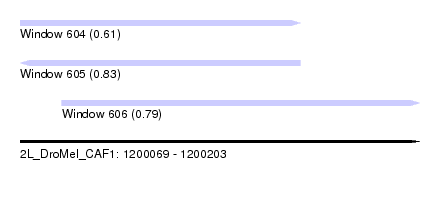
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,200,069 – 1,200,203 |
| Length | 134 |
| Max. P | 0.832038 |

| Location | 1,200,069 – 1,200,163 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.00 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1200069 94 + 22407834 UAUUGAGCGAAGUGUUGUUGUGGGAAAACCCGAAACUUACUUAUCAACCGAACUGGGCUCGUCUUAUCUGAAAUUUAGAUAAGAUUACAACGUA ....((((.(((((..(((.((((....)))).))).))))).....((.....))))))(((((((((((...)))))))))))......... ( -26.70) >DroSim_CAF1 17134 92 + 1 UAUUGAGCGAAGUGUUGUUGUGGGAAAACCUGAAA--UACUUAUCAGGCGAACUGGGCUCGUCUUAUCUGGAAUUUAGAUAAGAUUUCAACGUA ....((((.(((((((....((((....)))).))--))))).((((.....))))))))((((((((((.....))))))))))......... ( -26.00) >DroEre_CAF1 16970 86 + 1 UACUGGGCGUAGUGUUGUUGUGGGAAAGUCUGAGG--AACUUAUCAGCUGAACU------GUCUUAUCUGGCAUUUAGAUAAGAUUUCAACGUU .....((((((((...((((((((....((....)--).)))).))))...)))------((((((((((.....))))))))))....))))) ( -19.00) >consensus UAUUGAGCGAAGUGUUGUUGUGGGAAAACCUGAAA__UACUUAUCAGCCGAACUGGGCUCGUCUUAUCUGGAAUUUAGAUAAGAUUUCAACGUA ..((((...(((((......((((....)))).....))))).)))).............(((((((((((...)))))))))))......... (-16.66 = -16.00 + -0.66)

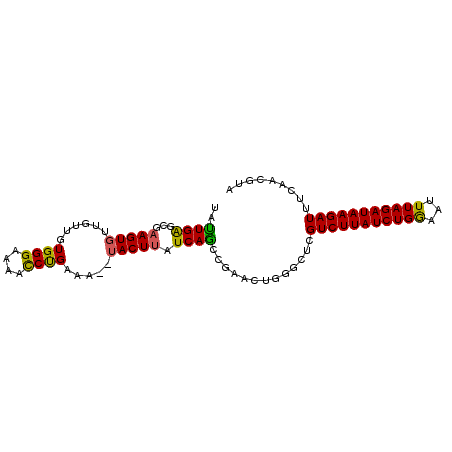
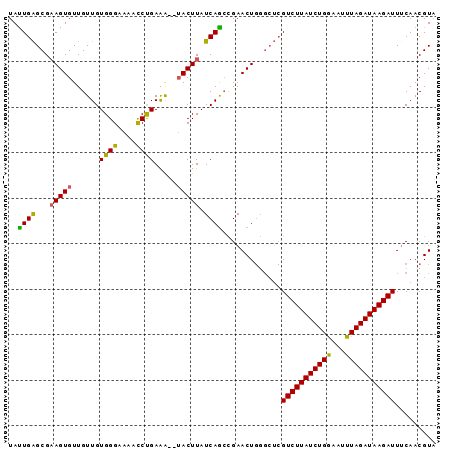
| Location | 1,200,069 – 1,200,163 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -19.03 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1200069 94 - 22407834 UACGUUGUAAUCUUAUCUAAAUUUCAGAUAAGACGAGCCCAGUUCGGUUGAUAAGUAAGUUUCGGGUUUUCCCACAACAACACUUCGCUCAAUA ..........((((((((.......)))))))).((((..(((...((((....(.......)(((....))).))))...)))..)))).... ( -21.60) >DroSim_CAF1 17134 92 - 1 UACGUUGAAAUCUUAUCUAAAUUCCAGAUAAGACGAGCCCAGUUCGCCUGAUAAGUA--UUUCAGGUUUUCCCACAACAACACUUCGCUCAAUA ...((((...((((((((.......)))))))).((((...))))((((((......--..)))))).......))))................ ( -20.40) >DroEre_CAF1 16970 86 - 1 AACGUUGAAAUCUUAUCUAAAUGCCAGAUAAGAC------AGUUCAGCUGAUAAGUU--CCUCAGACUUUCCCACAACAACACUACGCCCAGUA ...((((((.((((((((.......)))))))).------..)))))).((.(((((--.....)))))))....................... ( -15.10) >consensus UACGUUGAAAUCUUAUCUAAAUUCCAGAUAAGACGAGCCCAGUUCGGCUGAUAAGUA__UUUCAGGUUUUCCCACAACAACACUUCGCUCAAUA ...((((...((((((((.......))))))))..............((((..........)))).........))))................ (-12.77 = -12.33 + -0.44)

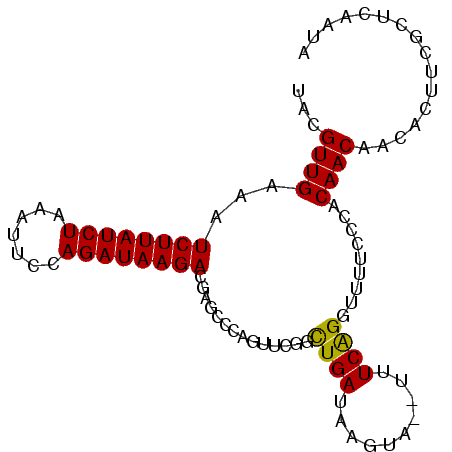
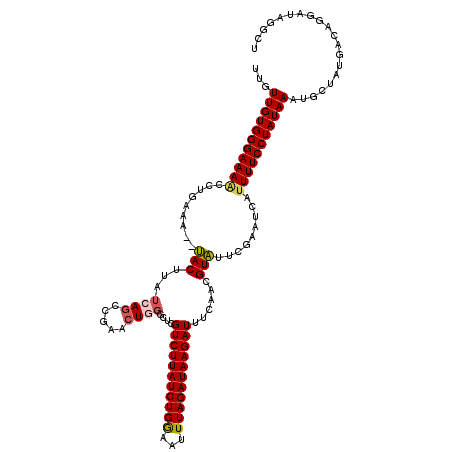
| Location | 1,200,083 – 1,200,203 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -19.31 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1200083 120 + 22407834 UUGUUGUGGGAAAACCCGAAACUUACUUAUCAACCGAACUGGGCUCGUCUUAUCUGAAAUUUAGAUAAGAUUACAACGUAUUCGAAUCAUUUUCCUAUAAAUGGUUUGACAGGAUAGGCU ..(((.((((....)))).)))...((((((..((.....))....(((((((((((...)))))))))))..........(((((((((((......)))))))))))...)))))).. ( -31.00) >DroSim_CAF1 17148 118 + 1 UUGUUGUGGGAAAACCUGAAA--UACUUAUCAGGCGAACUGGGCUCGUCUUAUCUGGAAUUUAGAUAAGAUUUCAACGUAUUCGAGCCAUUUUCCUAUAAAUGCUAUGAGAGGAUAGGCU ...((((((((((((((((..--......))))).......(((((((((((((((.....)))))))))............)))))).)))))))))))...((((......))))... ( -35.20) >DroEre_CAF1 16984 112 + 1 UUGUUGUGGGAAAGUCUGAGG--AACUUAUCAGCUGAACU------GUCUUAUCUGGCAUUUAGAUAAGAUUUCAACGUUUUCGAAUCUUUUUCCUAUAAAUGGAAUGACAGAAUAGGGU ...(((((((((((.((((..--......)))))((((..------((((((((((.....))))))))))))))...............)))))))))).................... ( -24.50) >DroYak_CAF1 16981 118 + 1 UUGUUGUGGGAAAUUCUGAAA--UACUUAUAAGCCCAACUGGGCUCGUCUUAUCUGGAAUUUAGAUAAGAUUUCAGCGUAUUCCAAUCAUUUUCCUAUAAAUGCUAUGACAGCAGAAGGU (((((((((((.((.(((((.--........(((((....))))).((((((((((.....))))))))))))))).)).))))...(((((......))))).....)))))))..... ( -34.30) >consensus UUGUUGUGGGAAAACCUGAAA__UACUUAUCAGCCGAACUGGGCUCGUCUUAUCUGGAAUUUAGAUAAGAUUUCAACGUAUUCGAAUCAUUUUCCUAUAAAUGCUAUGACAGGAUAGGCU ...(((((((((((.........(((...((((.....))))....(((((((((((...)))))))))))......))).........))))))))))).................... (-19.31 = -19.87 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:33 2006