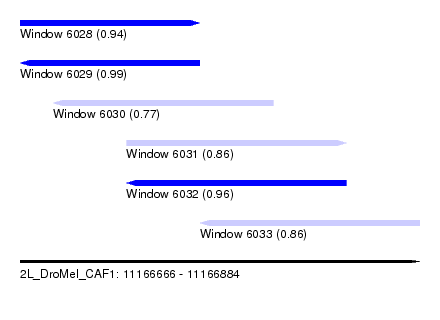
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,166,666 – 11,166,884 |
| Length | 218 |
| Max. P | 0.986277 |

| Location | 11,166,666 – 11,166,764 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.937976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11166666 98 + 22407834 GGAUGUUUGA----------UU-GUUU-GUGGCACCUGUGUGAAUAGUUUGUUGUAUCCACUCUGGUAGGAGGGGUAUCCAAGGCCAAAUCGAGACUCUCGAUGUUGGCC (((((..(..----------.(-(..(-(..(((.((((....))))..)))..))..))((((....)))))..)))))..((((((((((((...)))))).)))))) ( -33.10) >DroVir_CAF1 2809 102 + 1 ----GUUAUAGAAAAUAUAU---AUUU-UUUGCACCUGAGUGAAGAGUUUAUUGUAUCCGCUUUGGUAUGAUGUAUAUCCAAGGCCAAAUCGAGACUCUCGAUUAUGGCC ----..........((((((---(((.-.....(((.(((((................))))).)))..)))))))))....((((((((((((...))))))).))))) ( -25.69) >DroPse_CAF1 5122 105 + 1 AAAAGUUAUUCAAAAG----UUGUGUU-UUUGCACCUGUGUGAAGAGCUUGUUGUAUCCACUCUGGUAAGACGUGUAUCCAAGGCCAAAUCGAGACUCUCGAUGUUGGCC ..(((((.((((..((----..((((.-...))))))...)))).))))).(((.((.((((((....))).))).)).)))((((((((((((...)))))).)))))) ( -30.40) >DroWil_CAF1 4115 105 + 1 AAACAUAAUA-AAUAA----UACUUUGAUAUGCACCUGUGUAAAUAAUUUGUUAUAUCCACUUUGAUAUGAUGUAUAUCCAAGGCCAAAUCGAGAUUCUCGAUGUUGGCC ..........-.....----..(((.(((((((...(((....)))....((((((((......))))))))))))))).)))(((((((((((...)))))).))))). ( -26.50) >DroMoj_CAF1 3055 100 + 1 ----GUUAUAUUA--UUUGA---ACAU-UCUGCACCUGUGUGAAUAGCUUAUUGUAUCCGCUUUGGUAUGAUGUAUAUCCAAGGCCAAAUCGAGAUUCUCGAUGUUGGCC ----.........--.....---((((-(.(((....(((.((...((.....)).)))))....))).)))))........((((((((((((...)))))).)))))) ( -24.10) >DroAna_CAF1 3160 104 + 1 UGAUUUCUUA-AAUAA----UUCGUUG-UCAGCACCUGUGUGAAUAGUUUGUUGUAUCCACUCUGAUAGGAGGGGUAUCCAAGGCCAAAUCGAGACUCUCGAUGAUGGCC ..........-.....----.......-.(((((.((((....))))..)))))(((((.((((....))))))))).....(((((.((((((...))))))..))))) ( -28.90) >consensus __A_GUUAUA_AA_AA____UU_AUUU_UUUGCACCUGUGUGAAUAGUUUGUUGUAUCCACUCUGGUAUGAUGUAUAUCCAAGGCCAAAUCGAGACUCUCGAUGUUGGCC ...............................(((....)))........(((..((((......))))..))).........((((((((((((...)))))).)))))) (-16.30 = -16.72 + 0.42)

| Location | 11,166,666 – 11,166,764 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.09 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11166666 98 - 22407834 GGCCAACAUCGAGAGUCUCGAUUUGGCCUUGGAUACCCCUCCUACCAGAGUGGAUACAACAAACUAUUCACACAGGUGCCAC-AAAC-AA----------UCAAACAUCC ((((((.((((((...)))))))))))).(((.((((..((......))(((((((........)))))))...))))))).-....-..----------.......... ( -27.60) >DroVir_CAF1 2809 102 - 1 GGCCAUAAUCGAGAGUCUCGAUUUGGCCUUGGAUAUACAUCAUACCAAAGCGGAUACAAUAAACUCUUCACUCAGGUGCAAA-AAAU---AUAUAUUUUCUAUAAC---- (((((.(((((((...))))))))))))((((((....))).((((..((.(((............))).))..))))))).-....---................---- ( -20.30) >DroPse_CAF1 5122 105 - 1 GGCCAACAUCGAGAGUCUCGAUUUGGCCUUGGAUACACGUCUUACCAGAGUGGAUACAACAAGCUCUUCACACAGGUGCAAA-AACACAA----CUUUUGAAUAACUUUU ((((((.((((((...))))))))))))(((....(((.(((....))))))....))).......((((.....(((....-..)))..----....))))........ ( -23.50) >DroWil_CAF1 4115 105 - 1 GGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAUCAUAUCAAAGUGGAUAUAACAAAUUAUUUACACAGGUGCAUAUCAAAGUA----UUAUU-UAUUAUGUUU ((((((.((((((...))))))))))))((((((((.....)))))...(((((((........))))))).)))..(((((..(((...----...))-)..))))).. ( -26.00) >DroMoj_CAF1 3055 100 - 1 GGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAUCAUACCAAAGCGGAUACAAUAAGCUAUUCACACAGGUGCAGA-AUGU---UCAAA--UAAUAUAAC---- ((((((.((((((...))))))))))))((((((((...((.((((...(.(((((........))))).)...))))..))-))))---)))).--.........---- ( -25.70) >DroAna_CAF1 3160 104 - 1 GGCCAUCAUCGAGAGUCUCGAUUUGGCCUUGGAUACCCCUCCUAUCAGAGUGGAUACAACAAACUAUUCACACAGGUGCUGA-CAACGAA----UUAUU-UAAGAAAUCA (((((..((((((...)))))).)))))(..(.((((..((......))(((((((........)))))))...)))))..)-.......----.....-.......... ( -24.40) >consensus GGCCAACAUCGAGAGUCUCGAUUUGGCCUUGGAUACACAUCAUACCAAAGUGGAUACAACAAACUAUUCACACAGGUGCAAA_AAAA_AA____AU_UU_UAUAAC_U__ ((((((.((((((...))))))))))))..............((((...(((((((........)))))))...))))................................ (-20.28 = -20.92 + 0.64)
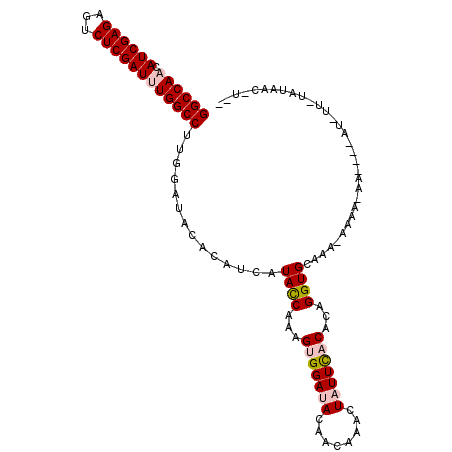
| Location | 11,166,684 – 11,166,804 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.97 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11166684 120 - 22407834 GCAGCGCGCCUCUUCCUCGGAUGCACCCUCCGUUCAGGGUGGCCAACAUCGAGAGUCUCGAUUUGGCCUUGGAUACCCCUCCUACCAGAGUGGAUACAACAAACUAUUCACACAGGUGCC .....((((((.......((....(((((......)))))((((((.((((((...))))))))))))..(((......)))..))...(((((((........)))))))..)))))). ( -40.90) >DroVir_CAF1 2831 120 - 1 GCAGCGUGCAUCAUCCUCAGAUGCUCCCUCAAUUCAGGGCGGCCAUAAUCGAGAGUCUCGAUUUGGCCUUGGAUAUACAUCAUACCAAAGCGGAUACAAUAAACUCUUCACUCAGGUGCA (((.((.(((((.......))))).)(((......)))(((((((.(((((((...))))))))))))((((.(((.....))))))).)).......................).))). ( -33.70) >DroPse_CAF1 5147 120 - 1 GCAGCGCGCAUCAUCCUCGGAUGCUCCCUCCGUUCAGGGUGGCCAACAUCGAGAGUCUCGAUUUGGCCUUGGAUACACGUCUUACCAGAGUGGAUACAACAAGCUCUUCACACAGGUGCA ...(((((((((.......))))).......((..(((((((((((.((((((...))))))))))))(((....(((.(((....))))))....)))...)))))..))....)))). ( -38.40) >DroGri_CAF1 4986 120 - 1 GCAGCGUGCAUCAUCCUCAGAUGCUCCCUCAAUCCAGGGCGGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAACAUAUCAAAGCGGAUACAAUAAGCUAUUCACACAGGUGCA (((.((.(((((.......))))).)(((......)))((((((((.((((((...))))))))))))...(((((.....)))))...))(((((........))))).....).))). ( -35.20) >DroWil_CAF1 4140 120 - 1 GCAACGCGCAUCGUCCUCAGAUGCUCCCUCCGUUCAGGGAGGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAUCAUAUCAAAGUGGAUAUAACAAAUUAUUUACACAGGUGCA .......(((((((((........(((((......)))))((((((.((((((...))))))))))))..))))...............(((((((........)))))))...))))). ( -39.10) >DroMoj_CAF1 3075 120 - 1 GCAGCGAGCAUCCUCCUCAGAUGCUCCCUCCAUUCAGGGUGGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAUCAUACCAAAGCGGAUACAAUAAGCUAUUCACACAGGUGCA (((.((((((((.......)))))))..(((.........((((((.((((((...))))))))))))((((.(((.....)))))))...)))....................).))). ( -36.70) >consensus GCAGCGCGCAUCAUCCUCAGAUGCUCCCUCCAUUCAGGGUGGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAUCAUACCAAAGCGGAUACAACAAACUAUUCACACAGGUGCA (((.((((((((.......))))).((((......)))).((((((.((((((...))))))))))))((((.(((.....))))))).))(((((........))))).....).))). (-30.80 = -30.97 + 0.17)



| Location | 11,166,724 – 11,166,844 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -46.57 |
| Consensus MFE | -38.79 |
| Energy contribution | -39.10 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11166724 120 + 22407834 AGGGGUAUCCAAGGCCAAAUCGAGACUCUCGAUGUUGGCCACCCUGAACGGAGGGUGCAUCCGAGGAAGAGGCGCGCUGCACCAUGGGCCUGUCAGCCCGGUAGUCUUCCUCGAACGAAG .(((((((((..((((((((((((...)))))).))))))..((.....)).)))))).)))((((((((((.((...)).)).(((((......)))))....))))))))........ ( -53.60) >DroVir_CAF1 2871 120 + 1 AUGUAUAUCCAAGGCCAAAUCGAGACUCUCGAUUAUGGCCGCCCUGAAUUGAGGGAGCAUCUGAGGAUGAUGCACGCUGCACCAUGGGCCUGUCAGCCCGAUAAUCUUCUUCGAAUGAAG ............((((((((((((...))))))).))))).((((......))))..(((.((((((.(((((.....))....(((((......)))))...))).)))))).)))... ( -40.90) >DroGri_CAF1 5026 120 + 1 UUGUAUAUCCAAGGCCAAAUCGAGAUUCUCGAUGUUGGCCGCCCUGGAUUGAGGGAGCAUCUGAGGAUGAUGCACGCUGCACCAUGGGCCUGUCAGCCCGAUAAUCUUCUUCGAAUGAAG .......((((.((((((((((((...)))))).))))))....))))(((((((((.(((.....(((.(((.....))).)))((((......)))))))...)))))))))...... ( -42.10) >DroWil_CAF1 4180 120 + 1 AUGUAUAUCCAAGGCCAAAUCGAGAUUCUCGAUGUUGGCCUCCCUGAACGGAGGGAGCAUCUGAGGACGAUGCGCGUUGCACCAUGGGCCUAUCAGCCCUAUAGUCUUCUUCGAAUGAAG ((((........((((((((((((...)))))).))))))(((((......)))))))))..(((((((.(((.....))).)..((((......))))....))))))........... ( -41.10) >DroMoj_CAF1 3115 120 + 1 AUGUAUAUCCAAGGCCAAAUCGAGAUUCUCGAUGUUGGCCACCCUGAAUGGAGGGAGCAUCUGAGGAGGAUGCUCGCUGCACCAUGGGCCUGUCAGCCCGAUAAUCCUCUUCGAAUGAAG ............((((((((((((...)))))).)))))).((((......))))..(((.((((((((((((.....))....(((((......)))))...)))))))))).)))... ( -47.90) >DroAna_CAF1 3224 120 + 1 AGGGGUAUCCAAGGCCAAAUCGAGACUCUCGAUGAUGGCCGCCCUGAACGGAGGGUGCAUCCGAGGAGGAUGCGCGCUGCACCAUGGGCCUGUCAGCCCGGUAGUCUUCUUCGAAGGAAG ..((((......(((((.((((((...))))))..)))))((((.....(.((.((((((((.....)))))))).)).).....))))......)))).........((((....)))) ( -53.80) >consensus AUGUAUAUCCAAGGCCAAAUCGAGACUCUCGAUGUUGGCCGCCCUGAACGGAGGGAGCAUCUGAGGAGGAUGCGCGCUGCACCAUGGGCCUGUCAGCCCGAUAAUCUUCUUCGAAUGAAG ............((((((((((((...)))))).)))))).((((......))))..(((.((((((.(((((.....)).....((((......))))....))).)))))).)))... (-38.79 = -39.10 + 0.31)


| Location | 11,166,724 – 11,166,844 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -46.12 |
| Consensus MFE | -38.79 |
| Energy contribution | -39.15 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11166724 120 - 22407834 CUUCGUUCGAGGAAGACUACCGGGCUGACAGGCCCAUGGUGCAGCGCGCCUCUUCCUCGGAUGCACCCUCCGUUCAGGGUGGCCAACAUCGAGAGUCUCGAUUUGGCCUUGGAUACCCCU ...(((((((((((((.....(((((....)))))..(((((...)))))))))))))))))).(((((......)))))((((((.((((((...))))))))))))..((.....)). ( -57.50) >DroVir_CAF1 2871 120 - 1 CUUCAUUCGAAGAAGAUUAUCGGGCUGACAGGCCCAUGGUGCAGCGUGCAUCAUCCUCAGAUGCUCCCUCAAUUCAGGGCGGCCAUAAUCGAGAGUCUCGAUUUGGCCUUGGAUAUACAU ((((....)))).........(((((....)))))..(((((.....)))))((((.........((((......)))).(((((.(((((((...))))))))))))..))))...... ( -42.10) >DroGri_CAF1 5026 120 - 1 CUUCAUUCGAAGAAGAUUAUCGGGCUGACAGGCCCAUGGUGCAGCGUGCAUCAUCCUCAGAUGCUCCCUCAAUCCAGGGCGGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAA ((((....))))..((.(((((((((....)))))(((((((.....))))))).....)))).)).....((((((...((((((.((((((...))))))))))))))))))...... ( -44.00) >DroWil_CAF1 4180 120 - 1 CUUCAUUCGAAGAAGACUAUAGGGCUGAUAGGCCCAUGGUGCAACGCGCAUCGUCCUCAGAUGCUCCCUCCGUUCAGGGAGGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAU ....((((((.((.(((....(((((....)))))..(((((.....)))))))).))......(((((......)))))((((((.((((((...))))))))))))))))))...... ( -44.80) >DroMoj_CAF1 3115 120 - 1 CUUCAUUCGAAGAGGAUUAUCGGGCUGACAGGCCCAUGGUGCAGCGAGCAUCCUCCUCAGAUGCUCCCUCCAUUCAGGGUGGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAU ....((((((.(((((.....(((((....)))))..(((((.....))))).))))).......((((......)))).((((((.((((((...))))))))))))))))))...... ( -44.70) >DroAna_CAF1 3224 120 - 1 CUUCCUUCGAAGAAGACUACCGGGCUGACAGGCCCAUGGUGCAGCGCGCAUCCUCCUCGGAUGCACCCUCCGUUCAGGGCGGCCAUCAUCGAGAGUCUCGAUUUGGCCUUGGAUACCCCU ((((....)))).........(((((....)))))..(((((...((((((((.....))))))..(((......)))))(((((..((((((...)))))).)))))...).))))... ( -43.60) >consensus CUUCAUUCGAAGAAGACUAUCGGGCUGACAGGCCCAUGGUGCAGCGCGCAUCAUCCUCAGAUGCUCCCUCCAUUCAGGGCGGCCAACAUCGAGAAUCUCGAUUUGGCCUUGGAUAUACAU ((((....)))).........(((((....)))))..(((((.....))))).(((.........((((......)))).((((((.((((((...))))))))))))..)))....... (-38.79 = -39.15 + 0.36)

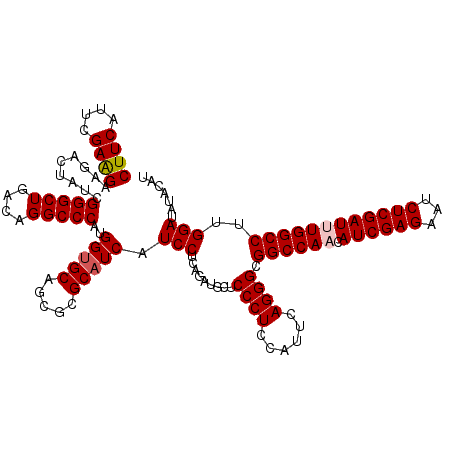

| Location | 11,166,764 – 11,166,884 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -42.36 |
| Consensus MFE | -30.07 |
| Energy contribution | -30.68 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11166764 120 - 22407834 UUGUCCUCGCACAGGAUCGUCGGAAACAUCGCCAAGAUAUCUUCGUUCGAGGAAGACUACCGGGCUGACAGGCCCAUGGUGCAGCGCGCCUCUUCCUCGGAUGCACCCUCCGUUCAGGGU ..(((((.....)))))((..(....)..))............(((((((((((((.....(((((....)))))..(((((...)))))))))))))))))).(((((......))))) ( -46.60) >DroVir_CAF1 2911 120 - 1 UUGUCGUUGUGCCGUUGUGUUGGAAACAUCAUCAAAAUAUCUUCAUUCGAAGAAGAUUAUCGGGCUGACAGGCCCAUGGUGCAGCGUGCAUCAUCCUCAGAUGCUCCCUCAAUUCAGGGC ....(((((..((((.(((.((....)).))).......(((((....)))))........(((((....)))))))))..))))).(((((.......))))).((((......)))). ( -41.70) >DroPse_CAF1 5227 120 - 1 UCGCUUACGCACCCUGUCGCCGACAAUAUCAUCAAGAUAUCUUCAUUCGAAGAAGACUACAGGGCUGACAGGCCCAUGGUGCAGCGCGCAUCAUCCUCGGAUGCUCCCUCCGUUCAGGGU .((((...(((((..(((........((((.....))))(((((....))))).)))....(((((....)))))..))))))))).(((((.......))))).((((......)))). ( -39.70) >DroWil_CAF1 4220 120 - 1 UUGUUGUUGCAACCUGUCGUCGAGUAGAUUGCCAAAAUAUCUUCAUUCGAAGAAGACUAUAGGGCUGAUAGGCCCAUGGUGCAACGCGCAUCGUCCUCAGAUGCUCCCUCCGUUCAGGGA ..((.((((((.((.(((.((((((((((.........))))..))))))....)))....(((((....)))))..))))))))))(((((.......)))))(((((......))))) ( -41.60) >DroMoj_CAF1 3155 118 - 1 --GUGUUUGGGCCAUUGGGUCGGUAACAUCAUCAAAAUAUCUUCAUUCGAAGAGGAUUAUCGGGCUGACAGGCCCAUGGUGCAGCGAGCAUCCUCCUCAGAUGCUCCCUCCAUUCAGGGU --(..((((((((.(..(.((((((..(((.........(((((....))))).))))))))).)..)..)))))).))..)...(((((((.......)))))))(((......))).. ( -38.40) >DroAna_CAF1 3264 120 - 1 CUGUUGUUGCACCGUGUCGUCACAAACACCAGCAAGAUAUCUUCCUUCGAAGAAGACUACCGGGCUGACAGGCCCAUGGUGCAGCGCGCAUCCUCCUCGGAUGCACCCUCCGUUCAGGGC ..((.((((((((((((((((..............))).(((((....))))).)))....(((((....)))))))))))))))))((((((.....)))))).((((......)))). ( -46.14) >consensus UUGUUGUUGCACCGUGUCGUCGGAAACAUCAUCAAAAUAUCUUCAUUCGAAGAAGACUACCGGGCUGACAGGCCCAUGGUGCAGCGCGCAUCAUCCUCAGAUGCUCCCUCCGUUCAGGGU ........(((((((........................(((((....)))))........(((((....)))))))))))).....(((((.......))))).((((......)))). (-30.07 = -30.68 + 0.61)
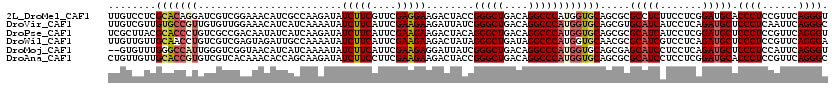

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:26 2006