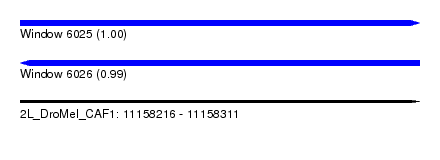
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,158,216 – 11,158,311 |
| Length | 95 |
| Max. P | 0.996007 |

| Location | 11,158,216 – 11,158,311 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -38.59 |
| Consensus MFE | -23.65 |
| Energy contribution | -22.66 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.996007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
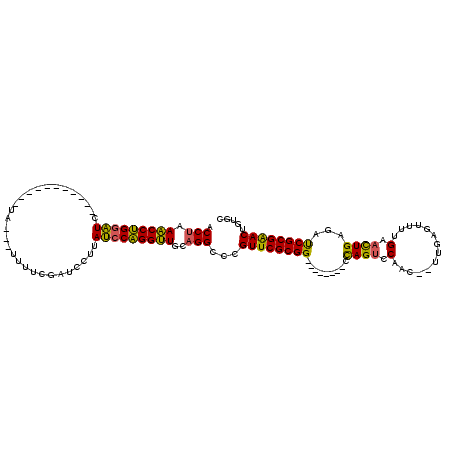
>2L_DroMel_CAF1 11158216 95 + 22407834 ACCUAAACCUGAAUC-----------GA---UUUUCGAUCCUUAUCCAGGUUGCAGGCCCGUUCGCGG--------CUAGUUCAAC--UUGAGUUUUGAACUGAGAUCGCGAACUGUGC .(((.((((((.(((-----------(.---....)))).......))))))..)))...((((((((--------(((((((((.--.......))))))).)).))))))))..... ( -31.40) >DroSec_CAF1 6539 95 + 1 ACCUAAACCUGGAUC-----------CA---UUGUCGAUCCUUAUCCAGGUUGCAGGUCCGUUCGCGG--------CUAGUUCAAC--UUGAGUUUUGAACUGAGGUCGCGAACUGUGC ((((.(((((((((.-----------..---............)))))))))..))))..((((((((--------(((((((((.--.......)))))))..))))))))))..... ( -38.26) >DroSim_CAF1 6575 95 + 1 ACCUAAACCUGGAUC-----------CA---UUGUCGAUCCUAAUCCAGGUUGCAGGUCCGUUCGCGG--------CUAGUUCAAC--UUGAGUUUGGAACUGAGAUCGCGAACUGUGC ((((.(((((((((.-----------..---............)))))))))..))))..((((((((--------(((((((.((--....))...))))).)).))))))))..... ( -34.36) >DroEre_CAF1 6525 95 + 1 ACCUAAACCUGGAUC-----------UG---UUUUCUAUCCUCAUCCAGGUUGCAGGCCUGUUCGCGG--------CCAGUCCAAC--UUGAGUUUUGAACUGAGUUCGCGAACUGUGC .(((.(((((((((.-----------.(---.........)..)))))))))..)))...((((((((--------(((((.(((.--.......))).)))).).))))))))..... ( -32.50) >DroYak_CAF1 6572 114 + 1 ACCCAAACCUGGAUUAUUUCGAUCCUUA---UUUUCGAUCCUUAUCCAGGUUGCAGGCCCGUUCGCGGUCUCCAGUCCAGUCCAAC--UGGAAUUUCGAACUGAGAUCGCGAACCGUGC .....(((((((((....((((......---...)))).....)))))))))(((.(...((((((((((((.(((((((.....)--))))........))))))))))))))).))) ( -45.10) >DroAna_CAF1 6266 106 + 1 ACCUAAGCCGGGGUCAU---GAUU--UGCUUUCUUGUAUCCUGACCCUGGCUGGAGGCUUGUUCGCGG--------GCAGUCCAACAUUGGAGCUGGGACUUGCCACCGCGGACUGUGC .(((.(((((((((((.---(((.--.((......))))).)))))))))))..)))...((((((((--------.(((((((....)))).)))((.....)).))))))))..... ( -49.90) >consensus ACCUAAACCUGGAUC___________UA___UUUUCGAUCCUUAUCCAGGUUGCAGGCCCGUUCGCGG________CCAGUCCAAC__UUGAGUUUUGAACUGAGAUCGCGAACUGUGC .(((.(((((((((.............................)))))))))..)))...((((((((.........((((.(..............).))))...))))))))..... (-23.65 = -22.66 + -0.99)


| Location | 11,158,216 – 11,158,311 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Mean single sequence MFE | -39.25 |
| Consensus MFE | -26.83 |
| Energy contribution | -26.08 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
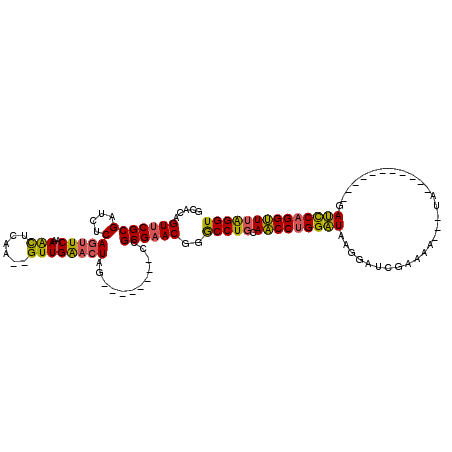
>2L_DroMel_CAF1 11158216 95 - 22407834 GCACAGUUCGCGAUCUCAGUUCAAAACUCAA--GUUGAACUAG--------CCGCGAACGGGCCUGCAACCUGGAUAAGGAUCGAAAA---UC-----------GAUUCAGGUUUAGGU ((...(((((((..((.(((((((.......--.)))))))))--------.)))))))..)).....((((((((..((((((....---.)-----------)))))..)))))))) ( -36.30) >DroSec_CAF1 6539 95 - 1 GCACAGUUCGCGACCUCAGUUCAAAACUCAA--GUUGAACUAG--------CCGCGAACGGACCUGCAACCUGGAUAAGGAUCGACAA---UG-----------GAUCCAGGUUUAGGU (((..(((((((..((.(((((((.......--.)))))))))--------.))))))).....))).((((((((..(((((.....---..-----------)))))..)))))))) ( -35.80) >DroSim_CAF1 6575 95 - 1 GCACAGUUCGCGAUCUCAGUUCCAAACUCAA--GUUGAACUAG--------CCGCGAACGGACCUGCAACCUGGAUUAGGAUCGACAA---UG-----------GAUCCAGGUUUAGGU .....(((((((..((.(((((..(((....--))))))))))--------.)))))))..(((((.((((((((((.(......)..---..-----------))))))))))))))) ( -35.30) >DroEre_CAF1 6525 95 - 1 GCACAGUUCGCGAACUCAGUUCAAAACUCAA--GUUGGACUGG--------CCGCGAACAGGCCUGCAACCUGGAUGAGGAUAGAAAA---CA-----------GAUCCAGGUUUAGGU .....(((((((...(((((((((.......--.)))))))))--------.)))))))..(((((.(((((((((..(.........---).-----------.)))))))))))))) ( -38.90) >DroYak_CAF1 6572 114 - 1 GCACGGUUCGCGAUCUCAGUUCGAAAUUCCA--GUUGGACUGGACUGGAGACCGCGAACGGGCCUGCAACCUGGAUAAGGAUCGAAAA---UAAGGAUCGAAAUAAUCCAGGUUUGGGU ...(.(((((((.((((((((((....(((.--...))).)))))).)))).))))))).)(((((.(((((((((.(...((((...---......))))..).)))))))))))))) ( -45.90) >DroAna_CAF1 6266 106 - 1 GCACAGUCCGCGGUGGCAAGUCCCAGCUCCAAUGUUGGACUGC--------CCGCGAACAAGCCUCCAGCCAGGGUCAGGAUACAAGAAAGCA--AAUC---AUGACCCCGGCUUAGGU .....((.(((((..(((.((((.(((......))))))))))--------))))).))..((((..((((.((((((.(((...........--.)))---.)))))).)))).)))) ( -43.30) >consensus GCACAGUUCGCGAUCUCAGUUCAAAACUCAA__GUUGAACUAG________CCGCGAACGGGCCUGCAACCUGGAUAAGGAUCGAAAA___UA___________GAUCCAGGUUUAGGU .....(((((((....)(((((..(((......))))))))............))))))..(((((.(((((((((.............................)))))))))))))) (-26.83 = -26.08 + -0.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:19 2006