
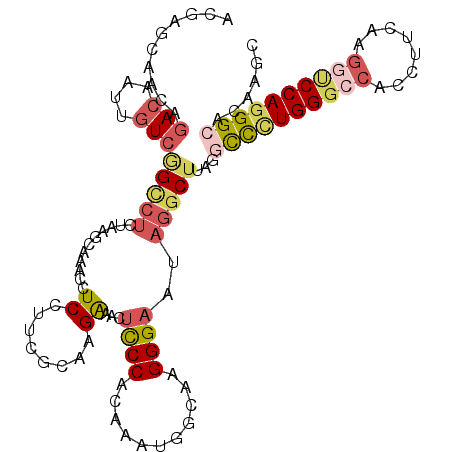
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,144,915 – 11,145,069 |
| Length | 154 |
| Max. P | 0.959381 |

| Location | 11,144,915 – 11,145,029 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -21.17 |
| Energy contribution | -22.32 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
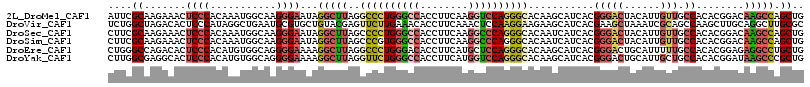
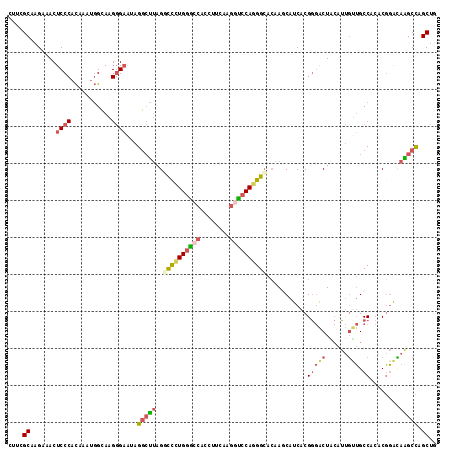
>2L_DroMel_CAF1 11144915 114 + 22407834 ACGUGCUGCAGACAAUUGUCGGUCUCUAAGCACACCUCAUUCGCAAGAAACUCCCACAAAUGGCAAGGGAAUAGGCUUAGGCCCUGGGCCACCUUCAAGGUCCAGGGCACAAGC ..(((((..((((........))))...)))))......(((....)))..((((...........))))....((((..((((((((((........))))))))))..)))) ( -45.60) >DroSec_CAF1 14115 114 + 1 ACGUGCUGCAGACAAUUGUCGGUCUCUAAGCACACCUCCUUCGCAAGAAACUCCCACAAAUGGCAAGGGAAUAGGCUUAGCCCCUGGGCCACCUUCAAGGCCCAGGGCACAAUC ..((((....(((....)))((...((((((........(((....)))..((((...........))))....))))))))((((((((........)))))))))))).... ( -42.60) >DroSim_CAF1 14140 114 + 1 ACGUGCUGCAGACAAUUGUCGGUCUCUACGCACACCUCCUUCGCAAGAAACUCCCACAAAUGGCAAGGGAAUAGGCUUAGCCCGUGGGCCACCUUCAAGGCCCAGGGCACAAUC ..((((((.((((........)))).)).)))).(((..(((....)))..((((...........))))..)))....((((.((((((........))))))))))...... ( -40.40) >DroEre_CAF1 14285 114 + 1 ACGAGCAACAGACAAUUGUCAGCCUCGAAGCAAACUUCCUGGGCCAGACACUCCCACAUGUGGCAGGGGAAAAGGCUUAGGCCCUGGGACACCUUCAUGCUCCAGGGCACAAGC .((((.....(((....)))...))))((((....(((((..((((.(..........).))))..)))))...))))..(((((((..((......))..)))))))...... ( -39.30) >DroYak_CAF1 14555 114 + 1 ACGAGCAACAGACAAUUGUCGGCCUCGAGGAAAACAUCCUUGGCGAGGCACUCCCACAUGUGGCAGGGGAAAAGGCUUAGGUUCUGGGCCACCUUCAUGGUCCAGGGCACAAGC ..((((....(((....))).((((((((((.....))))...))))))..((((.(........)))))....))))..(((((((((((......)))))))))))...... ( -43.40) >DroAna_CAF1 13843 114 + 1 GCGAGCAACACAAAGUAAUCGGCCUGAGGUCGAUUUUCGUUAAACAGGCACAUCCACAGGCAGACGGGGAGCUUACAGAGGCCUUGGACAAUCUUAAUGCACCAGGGCAGGAUC (..(((............((((((...))))))((..((((......((.(.......))).))))..)))))..)....(((((((.((.......))..)))))))...... ( -28.60) >consensus ACGAGCAACAGACAAUUGUCGGCCUCUAAGCAAACCUCCUUCGCAAGAAACUCCCACAAAUGGCAAGGGAAUAGGCUUAGGCCCUGGGCCACCUUCAAGGUCCAGGGCACAAGC ..........(((....)))(((((...........((........))...((((...........))))..)))))...((((((((((........))))))))))...... (-21.17 = -22.32 + 1.14)


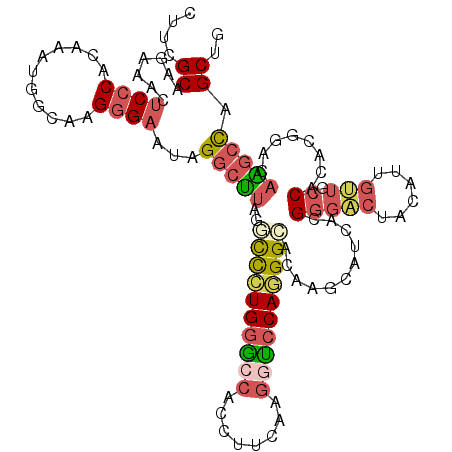
| Location | 11,144,953 – 11,145,069 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -40.87 |
| Consensus MFE | -25.15 |
| Energy contribution | -25.98 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11144953 116 + 22407834 AUUCGCAAGAAACUCCCACAAAUGGCAAGGGAAUAGGCUUAGGCCCUGGGCCACCUUCAAGGUCCAGGGCACAAGCAUCACGGGACUACAUUGUUGCCACACGGACAAGCCAGCUG .(((....)))...........((((((.((..(((((((..((((((((((........))))))))))..)))).((....)))))..)).))))))...((.....))..... ( -41.90) >DroVir_CAF1 15050 116 + 1 UCUGGCUAGACACUCCCAUAGGCUGAAUGCGUGCUGUACGAGUUCUUGAAACACCUUCAAACUCCAAGGAAGAAGCAUCACGAAGCUAAAUCGCAGCCAAGCUUGCAGGCUUGCGC ....((..............(((((.((((...((....(((((..((((.....)))))))))......))..))))..(((.......))))))))(((((....))))))).. ( -24.80) >DroSec_CAF1 14153 116 + 1 CUUCGCAAGAAACUCCCACAAAUGGCAAGGGAAUAGGCUUAGCCCCUGGGCCACCUUCAAGGCCCAGGGCACAAUCAUCACGGGACUACAUUGUUGCCACACGGACAAGCCAGCUG .(((....)))..((((...........))))...(((((..((.(((((((........)))))))((((((((..((....))....)))).))))....))..)))))..... ( -40.10) >DroSim_CAF1 14178 116 + 1 CUUCGCAAGAAACUCCCACAAAUGGCAAGGGAAUAGGCUUAGCCCGUGGGCCACCUUCAAGGCCCAGGGCACAAUCAUCACGGGACUACAUUGUUGCCACACGGACAAGCCAGCUG .(((....)))..((((...........))))...(((((.((((.((((((........))))))))))...........(((((......))).))........)))))..... ( -39.70) >DroEre_CAF1 14323 116 + 1 CUGGGCCAGACACUCCCACAUGUGGCAGGGGAAAAGGCUUAGGCCCUGGGACACCUUCAUGCUCCAGGGCACAAGCAUCACGGGACUGCAUUUUUGCCACACGGAGAGGCCUGCUG ..(((((.....((((....(((((((((((...((((((..(((((((..((......))..)))))))..)))).((....))))...))))))))))).)))).))))).... ( -48.00) >DroYak_CAF1 14593 116 + 1 CUUGGCGAGGCACUCCCACAUGUGGCAGGGGAAAAGGCUUAGGUUCUGGGCCACCUUCAUGGUCCAGGGCACAAGCAUCACGGGACUGCAUUGCUGCCACACGGAUAAGCCCGCUG ...((((.(((..(((....((((((((.((...((((((..(((((((((((......)))))))))))..)))).((....))))...)).)))))))).)))...))))))). ( -50.70) >consensus CUUCGCAAGAAACUCCCACAAAUGGCAAGGGAAUAGGCUUAGGCCCUGGGCCACCUUCAAGGUCCAGGGCACAAGCAUCACGGGACUACAUUGUUGCCACACGGACAAGCCAGCUG ....((.......((((...........))))...(((((..((((((((((........))))))))))...........(((((......))).))........))))).)).. (-25.15 = -25.98 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:14 2006