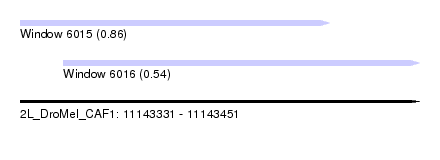
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,143,331 – 11,143,451 |
| Length | 120 |
| Max. P | 0.857303 |

| Location | 11,143,331 – 11,143,424 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11143331 93 + 22407834 UACUCACCUGAAUGGCACGGGCUACAUCCGAGCCCCAAAUCGGUUUCGAGCUGACUACUAUGGAGAAUUUAUUCCAAUGGAAGGCGUUCAGCA .......(((((((.(..(((((.......)))))....(((((.....)))))...((((((((......)))).))))..).))))))).. ( -26.40) >DroPse_CAF1 13498 93 + 1 CACUCACCUGUAUGGCCCGUGCCACAUCUGAGCCCCAAAUGGGCUUGGAGCUCACUACGAUGGCAAACUUUUGCCAGUGGAAGGCGCUCAGCA .......(((...(((((...((((.((..(((((.....)))))..))...........((((((....))))))))))..)).)))))).. ( -35.20) >DroEre_CAF1 12547 93 + 1 CACUCACUUGAAUGGCGCGGGCCACAUCCGAGCCCCAAAUCGGCUUGGAGCUGACUACUAUGGAGAACUUAUUCCAAUGAAAGGCGUUCAGUA .....(((.(((((.((..(..((..((((((((.......))))))))..)).)..)..(((((......)))))......).)))))))). ( -26.60) >DroMoj_CAF1 12953 93 + 1 UGCUUACCUGUAUAGCGCUGGCCACAUCGGAGCCCCAAAUGGGCUUCGAGCUAACCACAAUGGCGAAUUUUUUCCAAUCAUAGGCAUUUAAUA .(((.........)))....(((...(((((((((.....)))))))))((((.......))))..................)))........ ( -25.60) >DroAna_CAF1 12319 92 + 1 -CCUCACCUGAAUGGCCCGGGCUACGUCAGAACCAAAGAUUGGUUUGGAACUGACAACGAUGGAAAACUUUUCCCAAUGAAAAGCACUCAGCA -......((((.((((....)))).(((((..(((((......)))))..)))))............((((((.....))))))...)))).. ( -21.40) >DroPer_CAF1 13636 93 + 1 CACUCACCUGUAUGGCCCGUGCCACAUCUGAGCCCCAAAUGGGCUUGGAGCUCACUACGAUGGCAAACUUUUGCCAGUGGAAAGCGCUCAGCA .......(((..((((....))))..((..(((((.....)))))..))(((..((((..((((((....))))))))))..)))...))).. ( -32.90) >consensus CACUCACCUGAAUGGCCCGGGCCACAUCCGAGCCCCAAAUGGGCUUGGAGCUGACUACGAUGGAAAACUUUUUCCAAUGAAAGGCGCUCAGCA ............((((....))))..(((((((((.....)))))))))(((((......((((((....))))))(((.....)))))))). (-20.09 = -20.32 + 0.22)

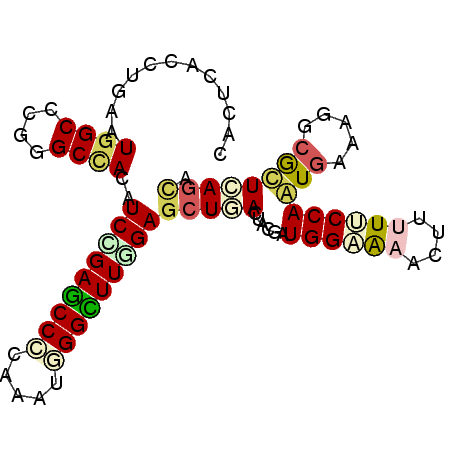
| Location | 11,143,344 – 11,143,451 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
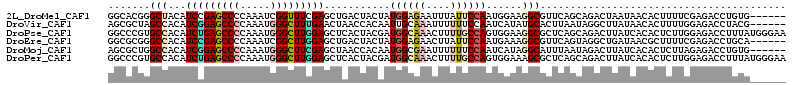
>2L_DroMel_CAF1 11143344 107 + 22407834 GGCACGGGCUACAUCCGAGCCCCAAAUCGGUUUCGAGCUGACUACUAUGGAGAAUUUAUUCCAAUGGAAGGCGUUCAGCAGACUAAUAACACUUUUCGAGACCUGUG------ ..((((((((.......)))))......(((((((((((((((.((((((((......)))).)))).))....))))).(........).....)))))))).)))------ ( -30.90) >DroVir_CAF1 14066 107 + 1 AGCGCUAGCCACAUCGGAGCCCCAAAUGGGCUUUGAACUAACCACAAUUGCAAAUUUUUUCCAAUCAUAUGCACUUAAUAGGCUUAUAACACUUUUGGAGACCUACG------ .((....))....(((((((((.....))))))))).....................(((((((..(((.((.((....)))).))).......)))))))......------ ( -21.20) >DroPse_CAF1 13511 113 + 1 GGCCCGUGCCACAUCUGAGCCCCAAAUGGGCUUGGAGCUCACUACGAUGGCAAACUUUUGCCAGUGGAAGGCGCUCAGCAGACUUAUCACACUCUUGGAGACCUUUAUGGGAA ..((((((((((.((..(((((.....)))))..))...........((((((....))))))))))((((..(((...(((..........)))..))).)))))))))).. ( -39.90) >DroEre_CAF1 12560 107 + 1 GGCGCGGGCCACAUCCGAGCCCCAAAUCGGCUUGGAGCUGACUACUAUGGAGAACUUAUUCCAAUGAAAGGCGUUCAGUAGGCUGAUAACGCUUUUCGAGACCUGCA------ ...(((((.((..((((((((.......))))))))..))....((.(((((......))))).((((((((((((((....))))..)))))))))))).))))).------ ( -41.40) >DroMoj_CAF1 12966 107 + 1 AGCGCUGGCCACAUCGGAGCCCCAAAUGGGCUUCGAGCUAACCACAAUGGCGAAUUUUUUCCAAUCAUAGGCAUUUAAUAGACUUAUCACACUCUUAGAGACCUGUG------ ..((((((.....(((((((((.....))))))))).....)).....)))).............((((((..(((((.((..........)).)))))..))))))------ ( -29.30) >DroPer_CAF1 13649 113 + 1 GGCCCGUGCCACAUCUGAGCCCCAAAUGGGCUUGGAGCUCACUACGAUGGCAAACUUUUGCCAGUGGAAAGCGCUCAGCAGACUUAUCACACUCUUGGAGACCUUUAUGGGAA ..(((((.((((.((..(((((.....)))))..))...........((((((....))))))))))((((.((((...(((..........)))..))).)))))))))).. ( -38.00) >consensus GGCGCGGGCCACAUCCGAGCCCCAAAUGGGCUUGGAGCUAACUACAAUGGCAAACUUUUUCCAAUGAAAGGCGCUCAGCAGACUUAUAACACUCUUGGAGACCUGUA______ .......(((...(((((((((.....)))))))))...........((((((....))))))......)))......................................... (-18.23 = -18.78 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:09 2006