
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,113,765 – 11,113,899 |
| Length | 134 |
| Max. P | 0.706455 |

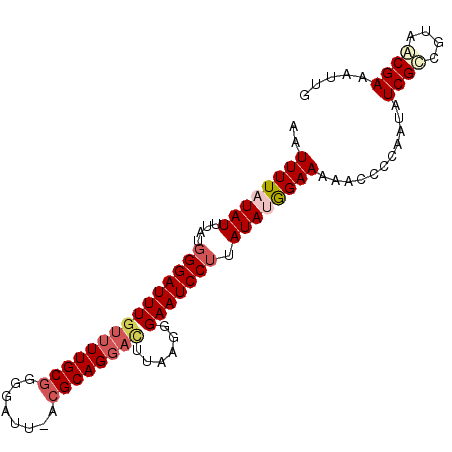
| Location | 11,113,765 – 11,113,860 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 87.70 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -17.18 |
| Energy contribution | -18.54 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11113765 95 + 22407834 AAUUUUAUAUUUAUGGGAUUUGUUUUGCGGGGAUUAACGCAGGACUUAGGGGAAUCCUUAUAUGGAAAAACCCCAAUGUCGCCGCAACGAAAAUG ..............(((((..((((((((........))))))))...((((..(((......)))....))))...))).))............ ( -24.40) >DroSec_CAF1 6674 94 + 1 AAUUUUAUAUUUAUGGGAUUUGUUUUGCGGGGAUU-ACGCAGGACUUAAGGGAAUCCUUAUAUGGAAAAACCCCAAUGUCGCUGUAGCGAAAUUG ..((((((((....(((((((((((((((......-.))))))))......))))))).))))))))......((((.((((....)))).)))) ( -26.70) >DroSim_CAF1 6920 86 + 1 AAUUUUAUAUUUAUGGGAUUUGUUUUGCGGGGAUU-ACGCAG--------GGAAUCCUUAUAUGGAAAAACCCCAAUAUCGCCGUAACGAAAAUG .(((((....((((((((((((...((((......-.))))(--------((..(((......)))....)))))).))).))))))..))))). ( -19.80) >DroEre_CAF1 6803 92 + 1 GAUUUUAUAUUUAUGGGAUUUGUUUUGCGGGGAUU-AGGCAGGAUUUAAGGGAAUCCGUAUAAGGAAAA-C-CAAAUAUCGUCGUAACGAAAUUG ..................(((((..(((((.(((.-.....((((((....))))))......((....-)-)....))).)))))))))).... ( -16.30) >DroYak_CAF1 6784 93 + 1 GAUUUUAUAUUUAUGGGAUUUGUUUUGCGGGGAUU-ACGCAGGACUUAAGGGAAUCCUUAUAAAGAAAA-CCCAAAUAUCGUCGUGACGAAAUUG .............((((....((((((((......-.)))))))).(((((....))))).........-))))(((.((((....)))).))). ( -23.40) >consensus AAUUUUAUAUUUAUGGGAUUUGUUUUGCGGGGAUU_ACGCAGGACUUAAGGGAAUCCUUAUAUGGAAAAACCCCAAUAUCGCCGUAACGAAAUUG ..((((((((....(((((((((((((((........))))))))......))))))).))))))))...........((((....))))..... (-17.18 = -18.54 + 1.36)


| Location | 11,113,765 – 11,113,860 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 87.70 |
| Mean single sequence MFE | -17.38 |
| Consensus MFE | -11.94 |
| Energy contribution | -12.58 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11113765 95 - 22407834 CAUUUUCGUUGCGGCGACAUUGGGGUUUUUCCAUAUAAGGAUUCCCCUAAGUCCUGCGUUAAUCCCCGCAAAACAAAUCCCAUAAAUAUAAAAUU ..........((((.(((...((((....(((......)))..))))...)))))))...................................... ( -20.80) >DroSec_CAF1 6674 94 - 1 CAAUUUCGCUACAGCGACAUUGGGGUUUUUCCAUAUAAGGAUUCCCUUAAGUCCUGCGU-AAUCCCCGCAAAACAAAUCCCAUAAAUAUAAAAUU .....((((....))))...((((((((.......(((((....))))).((..((((.-......))))..))))))))))............. ( -22.50) >DroSim_CAF1 6920 86 - 1 CAUUUUCGUUACGGCGAUAUUGGGGUUUUUCCAUAUAAGGAUUCC--------CUGCGU-AAUCCCCGCAAAACAAAUCCCAUAAAUAUAAAAUU .....((((....))))...((((((((.(((......)))....--------.((((.-......))))....))))))))............. ( -17.10) >DroEre_CAF1 6803 92 - 1 CAAUUUCGUUACGACGAUAUUUG-G-UUUUCCUUAUACGGAUUCCCUUAAAUCCUGCCU-AAUCCCCGCAAAACAAAUCCCAUAAAUAUAAAAUC (((..((((....))))...)))-(-((((........(((((......))))).((..-.......)))))))..................... ( -9.40) >DroYak_CAF1 6784 93 - 1 CAAUUUCGUCACGACGAUAUUUGGG-UUUUCUUUAUAAGGAUUCCCUUAAGUCCUGCGU-AAUCCCCGCAAAACAAAUCCCAUAAAUAUAAAAUC .(((.((((....)))).)))((((-.(((.....(((((....))))).((..((((.-......))))..))))).))))............. ( -17.10) >consensus CAAUUUCGUUACGGCGAUAUUGGGGUUUUUCCAUAUAAGGAUUCCCUUAAGUCCUGCGU_AAUCCCCGCAAAACAAAUCCCAUAAAUAUAAAAUU .....((((....))))...((((((((.(((......))).............((((........))))....))))))))............. (-11.94 = -12.58 + 0.64)

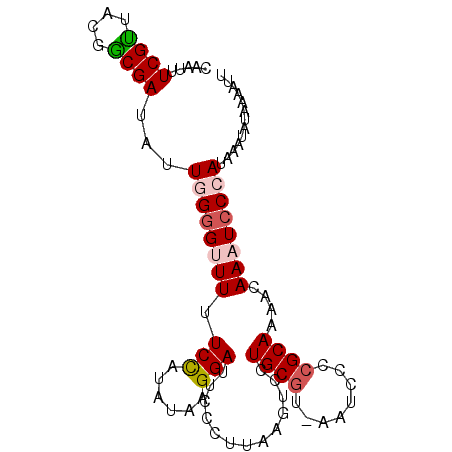

| Location | 11,113,781 – 11,113,899 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.31 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -10.56 |
| Energy contribution | -11.40 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11113781 118 - 22407834 AUGCAAAAGAUUCUAGGAAAUGUCUGCUCAAUUCCUAUUCAUUUUCGUUGCGGCGACAUUGGGGUUUUUCCAUAUAAGGAUUCCCCUAAGUCCUGCGUUAAUCCCCGCAAAACAAAUC .(((....((...((((((.((......)).)))))).)).........((((.(((...((((....(((......)))..))))...)))))))..........)))......... ( -31.70) >DroSec_CAF1 6690 103 - 1 AUCCAUAAGAUUC--------------UCAAAUCCUAUUCAAUUUCGCUACAGCGACAUUGGGGUUUUUCCAUAUAAGGAUUCCCUUAAGUCCUGCGU-AAUCCCCGCAAAACAAAUC .............--------------...((((((...((((.((((....)))).))))(((....))).....)))))).......((..((((.-......))))..))..... ( -20.60) >DroSim_CAF1 6936 95 - 1 AUCCAAAAGAUUC--------------UCAAUUCCUAUUCAUUUUCGUUACGGCGAUAUUGGGGUUUUUCCAUAUAAGGAUUCC--------CUGCGU-AAUCCCCGCAAAACAAAUC .............--------------............((...((((....))))...))(((....(((......)))..))--------)((((.-......))))......... ( -14.50) >DroEre_CAF1 6819 112 - 1 AUCCA---GAUUCUAGGCAAUGUCUGCUUAAUUUCUAUUCAAUUUCGUUACGACGAUAUUUG-G-UUUUCCUUAUACGGAUUCCCUUAAAUCCUGCCU-AAUCCCCGCAAAACAAAUC .....---(((..((((((...................((((..((((....))))...)))-)-............(((((......))))))))))-))))............... ( -17.10) >DroYak_CAF1 6800 96 - 1 A---------UUCCACGCAAUGUCU-----------ACUCAAUUUCGUCACGACGAUAUUUGGG-UUUUCUUUAUAAGGAUUCCCUUAAGUCCUGCGU-AAUCCCCGCAAAACAAAUC .---------....((((...(...-----------((((((..((((....))))...)))))-)...)......((((((......))))))))))-................... ( -19.70) >consensus AUCCA_AAGAUUC_A_G_AAUGUCU__UCAAUUCCUAUUCAAUUUCGUUACGGCGAUAUUGGGGUUUUUCCAUAUAAGGAUUCCCUUAAGUCCUGCGU_AAUCCCCGCAAAACAAAUC .......................................((((.((((....)))).))))(((....(((......)))..)))........((((........))))......... (-10.56 = -11.40 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:54 2006