
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,104,838 – 11,105,004 |
| Length | 166 |
| Max. P | 0.997164 |

| Location | 11,104,838 – 11,104,931 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -25.76 |
| Energy contribution | -24.77 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11104838 93 + 22407834 GUCCUCAUCGUCAUCAUUUAGGUGCAGGAGAUCAGCCUCAGGUGGCUCCUGAGGCAGC---GCCGCCGAUGACGAGGAUGUCACCCCCUGUCCACU .......((((((((.....(((((..(....).((((((((.....)))))))).))---)))...))))))))(((((........)))))... ( -39.40) >DroPse_CAF1 8983 87 + 1 GUCCUCGUCGUCGUCGUUCAAAUGCAGGAGAUCAGCCUCGGGGGGCUCUUGAGGUAAC---GACGACGACGAGGAGGAGGUCACUCC------ACC .((((((((((((((((((.....(((((.....((((....)))))))))..).)))---))))))))))))))((((....))))------... ( -48.40) >DroGri_CAF1 9045 88 + 1 AUCCUCGUCGUCGUCAUUCAAAUGCAGGAGAUCAGCCUCGGGCGGCUCUUGAGGUAUGUGCGCCGUUGAUGACGACGACGUCG-A---CAAU---- .((..((((((((((((.(((...((((((....(((...)))..)))))).(((......))).)))))))))))))))..)-)---....---- ( -37.90) >DroYak_CAF1 8000 93 + 1 GUCCUCAUCGUCAUCAUUUAGGUGCAGGAGAUCAGCCUCAGGUGGCUCCUGAGGCAGC---GCCGCCGACGACGAGGAUGUCACUCCCUGACCACU ((((((.(((((........(((((..(....).((((((((.....)))))))).))---)))...))))).))))))((((.....)))).... ( -40.80) >DroAna_CAF1 7595 90 + 1 GUCCUCCUCGUCAUCGUUUAGGUGCAGGAGAUCGGCUUCCGGCGGCUCCUGGGGCUGC---GACGUCGCCGACGAAGAGCCCACUCCACUUCC--- (..(((.(((((.......((.(.((((((.(((.(....).))))))))).).))((---(....))).))))).)))..)...........--- ( -32.00) >DroPer_CAF1 8941 87 + 1 GUCCUCGUCGUCGUCGUUCAAAUGCAGGAGAUCAGCCUCGGGGGGCUCUUGAGGUAAC---GACGACGACGAGGAGGAGGUCACUCC------ACC .((((((((((((((((((.....(((((.....((((....)))))))))..).)))---))))))))))))))((((....))))------... ( -48.40) >consensus GUCCUCGUCGUCAUCAUUCAAAUGCAGGAGAUCAGCCUCGGGCGGCUCCUGAGGCAGC___GACGACGACGACGAGGAGGUCACUCC____C_AC_ .(((((.((((((((.(((.......))))))..((((((((.....))))))))............))))).))))).................. (-25.76 = -24.77 + -1.00)

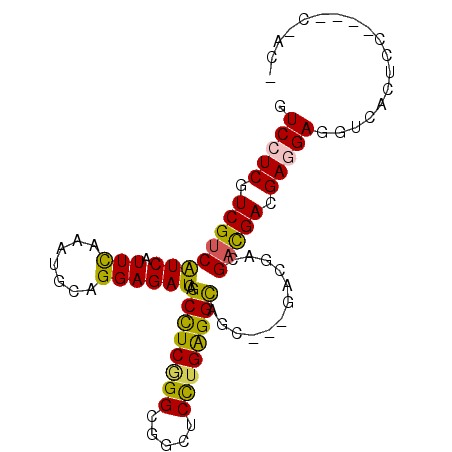
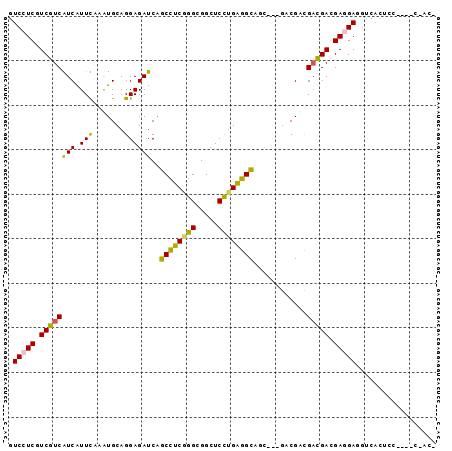
| Location | 11,104,838 – 11,104,931 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 74.59 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.41 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11104838 93 - 22407834 AGUGGACAGGGGGUGACAUCCUCGUCAUCGGCGGC---GCUGCCUCAGGAGCCACCUGAGGCUGAUCUCCUGCACCUAAAUGAUGACGAUGAGGAC ........((((....).)))(((((((((..((.---((.((((((((.....)))))))).........)).))....)))))))))....... ( -35.50) >DroPse_CAF1 8983 87 - 1 GGU------GGAGUGACCUCCUCCUCGUCGUCGUC---GUUACCUCAAGAGCCCCCCGAGGCUGAUCUCCUGCAUUUGAACGACGACGACGAGGAC ...------((((....))))((((((((((((((---((....(((((.((.....((((....))))..)).))))))))))))))))))))). ( -40.80) >DroGri_CAF1 9045 88 - 1 ----AUUG---U-CGACGUCGUCGUCAUCAACGGCGCACAUACCUCAAGAGCCGCCCGAGGCUGAUCUCCUGCAUUUGAAUGACGACGACGAGGAU ----...(---(-(..(((((((((((((((.(((...............)))((..((((....))))..))..))).))))))))))))..))) ( -32.16) >DroYak_CAF1 8000 93 - 1 AGUGGUCAGGGAGUGACAUCCUCGUCGUCGGCGGC---GCUGCCUCAGGAGCCACCUGAGGCUGAUCUCCUGCACCUAAAUGAUGACGAUGAGGAC ....((((.....)))).((((((((((((((((.---.(.((((((((.....)))))))).).....))))..(.....).)))))))))))). ( -40.70) >DroAna_CAF1 7595 90 - 1 ---GGAAGUGGAGUGGGCUCUUCGUCGGCGACGUC---GCAGCCCCAGGAGCCGCCGGAAGCCGAUCUCCUGCACCUAAACGAUGACGAGGAGGAC ---..............(((((((((((((....)---)).....((((((.((.(....).))..))))))...........))))))))))... ( -31.60) >DroPer_CAF1 8941 87 - 1 GGU------GGAGUGACCUCCUCCUCGUCGUCGUC---GUUACCUCAAGAGCCCCCCGAGGCUGAUCUCCUGCAUUUGAACGACGACGACGAGGAC ...------((((....))))((((((((((((((---((....(((((.((.....((((....))))..)).))))))))))))))))))))). ( -40.80) >consensus _GU_G____GGAGUGACCUCCUCGUCGUCGACGGC___GCUACCUCAAGAGCCACCCGAGGCUGAUCUCCUGCACCUAAACGACGACGACGAGGAC ..................(((((.((((((.((.....((.........((((......))))........)).......)).)))))).))))). (-15.61 = -16.41 + 0.81)



| Location | 11,104,909 – 11,105,004 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.00 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11104909 95 - 22407834 CAACAACAUUCCCUUGCCCACAAGUCUGCCCCACUGUUUGCAUCCCAACGUGCUACAAGC------AGCUGCGUCCGGCAGUGGACAGGGGGUGACAUCCU .......................(((.(((((.(((((..(..((..(((((((......------))).))))..))..)..)))))))))))))..... ( -32.40) >DroSec_CAF1 8191 95 - 1 CAACAACAUUCCCUUGCCCACAAGUCUGCCCCACUGUUUGCAUCCCAACGUGCUACAAGC------GGCUGUGUCCGGCAGUGGGCAGGGGGUGACAUCCU ......(((.(((((((((((.....(((((((((((((((((......))))...))))------)).)).)...))))))))))))))))))....... ( -35.00) >DroSim_CAF1 8449 95 - 1 CAACAACAUUCCCUUGCCCACAAGUCUGCCCCACUGUUUGCAUCCCAACGUGCUACAAGC------GGCUGCGUCCGGCAGUGGACAGGGGGUGACAUCAU .......................(((.(((((.(((((..(..((..(((((((......------))).))))..))..)..)))))))))))))..... ( -32.40) >DroEre_CAF1 8318 95 - 1 CAACAACAUUCCCUUGCCCACAAGUCUGCCCCAUUGCUUGCAUCCCAACGUGUUGCAAGC------GGCUGCGUCCGGCAGUGGACAGGGGGUGACAUCCU ......(((.((((((.((((......(((.....(((((((...........)))))))------)))(((.....))))))).)))))))))....... ( -33.00) >DroYak_CAF1 8071 95 - 1 CAACAACAUUCCUUUGCCCACUAGUCUGCCCCAAUGUUUGCAUCCUAACGUAUUACAAGC------GGCUGCAUCCGGCAGUGGUCAGGGAGUGACAUCCU ......((((((((.(.(((((.((((((.....((((((....(....).....)))))------)...)))...)))))))).)))))))))....... ( -25.30) >DroPer_CAF1 9012 95 - 1 CAACAAUAUACCGCUGCCCACCAGCCUGCCCCACUGUCUGCAUCCGAAUGUGCUGCAGACGAAGGCGGUUGGGGGCAGCGGU------GGAGUGACCUCCU .........((((((((((.((((((.(((....((((((((.((....).).))))))))..)))))))))))))))))))------((((....)))). ( -52.80) >consensus CAACAACAUUCCCUUGCCCACAAGUCUGCCCCACUGUUUGCAUCCCAACGUGCUACAAGC______GGCUGCGUCCGGCAGUGGACAGGGGGUGACAUCCU ......((((((((.(.((((.............(((..((((......)))).)))..........((((....)))).)))).)))))))))....... (-22.05 = -22.00 + -0.05)

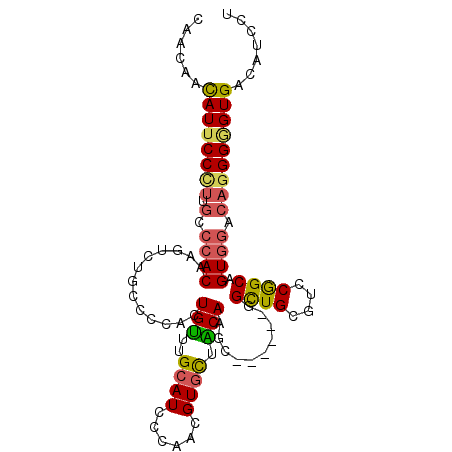
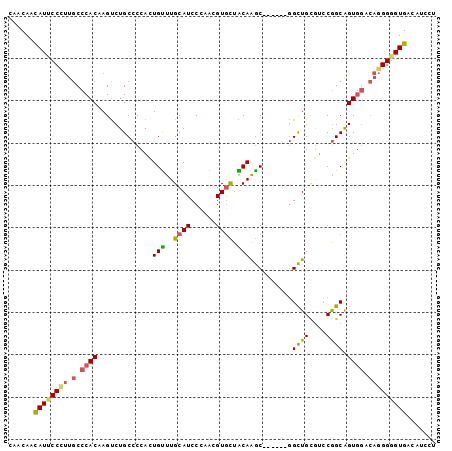
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:48 2006