
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,103,664 – 11,103,805 |
| Length | 141 |
| Max. P | 0.998594 |

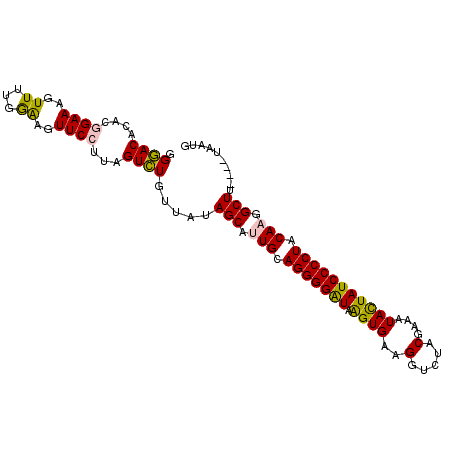
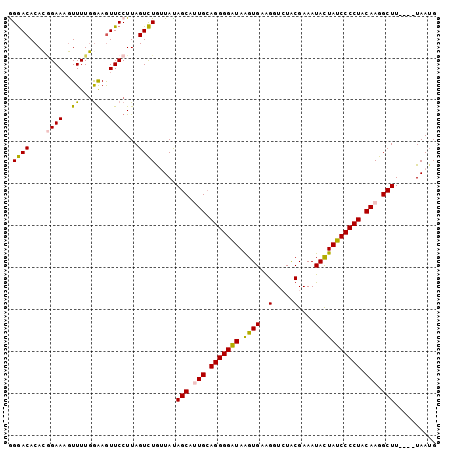
| Location | 11,103,664 – 11,103,766 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.94 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -20.10 |
| Energy contribution | -19.62 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11103664 102 + 22407834 GGAACACACGGAAUGUUUUGGAAGUUCCUUAGUUUGUUAUAGCAUUGCAGGGGAUAAGUGCGGAUCUACGAAAUACUAUCCCCUACAAGGCUUUAUUUAAUG (((((...((((....))))...)))))...(((...((.(((.(((.((((((((.(((((......))...))))))))))).))).))).))...))). ( -27.90) >DroSec_CAF1 6891 98 + 1 AGGACACACGGAAAGUUUUGGAAGUUCCUUAGUCUGUUAUAGCAUUGAAGGGGAUAAGUGAAGGUAUACGAAAUACUAUCCCCUACAAGGCUU----UAAUG .((((....((((..((...))..))))...))))((((.(((.(((.((((((((.(((..(.....)....))))))))))).))).))).----)))). ( -27.20) >DroSim_CAF1 7149 98 + 1 AGGACACACGGAAAGUUUUGGAAGUUCCUUAGUCUGUUAUAGCAUUGCAGGGGAUAAGUGAAGGUGUACGAAAUACUAUCCCCUACAAGGCUU----UAAUG .((((....((((..((...))..))))...))))((((.(((.(((.((((((((.(((..(.....)....))))))))))).))).))).----)))). ( -27.20) >DroEre_CAF1 7028 98 + 1 GGGACACAGCGAAAUUUUUCGAAAUUCCUUAGUCUAAUACAGCAAUGCAGGGGGUAGGUGUAGGUCUACGAAAUAUUAUCCCCUACAAGGCUU----UAAUG ((((.....((((....))))....))))........((.(((..((.(((((((((.((((....)))).....))))))))).))..))).----))... ( -25.10) >DroYak_CAF1 6767 98 + 1 GGGACACACAGAAAGUUUUGAGAGUUCCUUAGUCUAAUACAGCAAUGCAGGGGAUAGGUGUAGGUCUGCGAAAUAUUAUCCCCUACAAGGCUU----UAAAG (((((.(.((((....)))).).))))).........((.(((..((.(((((((((.((((....)))).....))))))))).))..))).----))... ( -27.80) >consensus GGGACACACGGAAAGUUUUGGAAGUUCCUUAGUCUGUUAUAGCAUUGCAGGGGAUAAGUGAAGGUCUACGAAAUACUAUCCCCUACAAGGCUU____UAAUG .((((....((((..((...))..))))...)))).....(((.(((.(((((((.((((..(.....)....))))))))))).))).))).......... (-20.10 = -19.62 + -0.48)

| Location | 11,103,664 – 11,103,766 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.94 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11103664 102 - 22407834 CAUUAAAUAAAGCCUUGUAGGGGAUAGUAUUUCGUAGAUCCGCACUUAUCCCCUGCAAUGCUAUAACAAACUAAGGAACUUCCAAAACAUUCCGUGUGUUCC ..........(((.(((((((((((((.....((......))...))))))))))))).)))............(((((..................))))) ( -26.67) >DroSec_CAF1 6891 98 - 1 CAUUA----AAGCCUUGUAGGGGAUAGUAUUUCGUAUACCUUCACUUAUCCCCUUCAAUGCUAUAACAGACUAAGGAACUUCCAAAACUUUCCGUGUGUCCU ..(((----.(((.(((.((((((((((((.....)))).......)))))))).))).))).)))..(((...((((...........))))....))).. ( -23.51) >DroSim_CAF1 7149 98 - 1 CAUUA----AAGCCUUGUAGGGGAUAGUAUUUCGUACACCUUCACUUAUCCCCUGCAAUGCUAUAACAGACUAAGGAACUUCCAAAACUUUCCGUGUGUCCU ..(((----.(((.((((((((((((((...............)).)))))))))))).))).)))..(((...((((...........))))....))).. ( -27.26) >DroEre_CAF1 7028 98 - 1 CAUUA----AAGCCUUGUAGGGGAUAAUAUUUCGUAGACCUACACCUACCCCCUGCAUUGCUGUAUUAGACUAAGGAAUUUCGAAAAAUUUCGCUGUGUCCC .....----.(((..((((((((..........((((........))))))))))))..)))......(((...(((((((....))))))).....))).. ( -24.50) >DroYak_CAF1 6767 98 - 1 CUUUA----AAGCCUUGUAGGGGAUAAUAUUUCGCAGACCUACACCUAUCCCCUGCAUUGCUGUAUUAGACUAAGGAACUCUCAAAACUUUCUGUGUGUCCC .....----.(((..(((((((((((....................)))))))))))..)))......(((..(((((..........)))))....))).. ( -23.85) >consensus CAUUA____AAGCCUUGUAGGGGAUAGUAUUUCGUAGACCUACACUUAUCCCCUGCAAUGCUAUAACAGACUAAGGAACUUCCAAAACUUUCCGUGUGUCCC ..........(((.(((((((((((((..................))))))))))))).)))......(((...((((...........))))....))).. (-21.99 = -22.95 + 0.96)

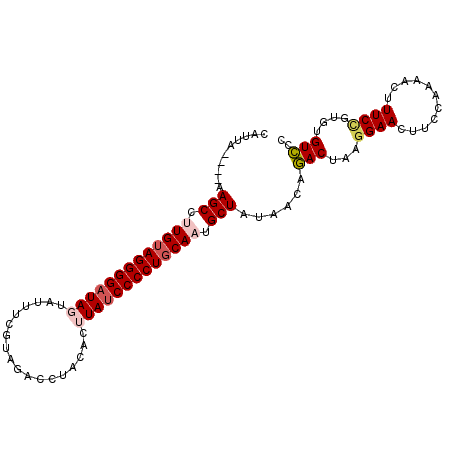
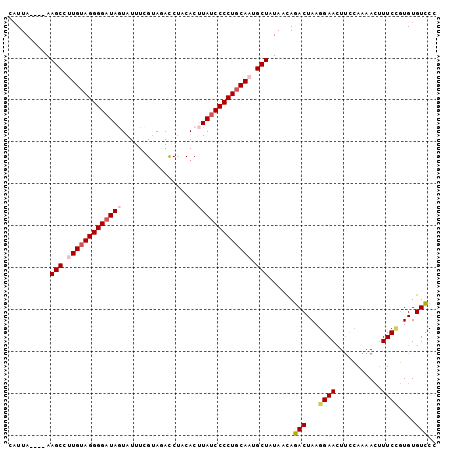
| Location | 11,103,686 – 11,103,805 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.36 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11103686 119 + 22407834 AGUUCCUUAGUUUGUUAUAGCAUUGCAGGGGAUAAGUGCGGAUCUACGAAAUACUAUCCCCUACAAGGCUUUAUUUAAUGACCAAGGCAUAUAAAACGACACAGAUUGAAAUUCUCAGA ......(((((((((((.(((.(((.((((((((.(((((......))...))))))))))).))).))).))(((((((.(....)))).)))).....))))))))).......... ( -27.50) >DroSec_CAF1 6913 115 + 1 AGUUCCUUAGUCUGUUAUAGCAUUGAAGGGGAUAAGUGAAGGUAUACGAAAUACUAUCCCCUACAAGGCUU----UAAUGACCAAAGCAUAUAAAACGACACAGAUUGAAAUACGCAGA .((...(((((((((.......(((.((((((((.(((..(.....)....))))))))))).))).((((----(.......)))))............)))))))))...))..... ( -28.20) >DroSim_CAF1 7171 115 + 1 AGUUCCUUAGUCUGUUAUAGCAUUGCAGGGGAUAAGUGAAGGUGUACGAAAUACUAUCCCCUACAAGGCUU----UAAUGACCAAAGCAUAUAAAACGACACAGAUUGAAAUACGCAGA .((...(((((((((.......(((.((((((((.(((..(.....)....))))))))))).))).((((----(.......)))))............)))))))))...))..... ( -28.20) >DroEre_CAF1 7050 115 + 1 AAUUCCUUAGUCUAAUACAGCAAUGCAGGGGGUAGGUGUAGGUCUACGAAAUAUUAUCCCCUACAAGGCUU----UAAUGACCAAGACAAAUUAAACGACACAGACUGAAAUACGCAAG ......(((((((.....(((..((.(((((((((.((((....)))).....))))))))).))..)))(----((((...........))))).......)))))))....(....) ( -26.50) >DroYak_CAF1 6789 115 + 1 AGUUCCUUAGUCUAAUACAGCAAUGCAGGGGAUAGGUGUAGGUCUGCGAAAUAUUAUCCCCUACAAGGCUU----UAAAGACCAAGGCAAUCAAAACGACACAAUUUGAAAUACGCAAG ....((((.((((..((.(((..((.(((((((((.((((....)))).....))))))))).))..))).----)).)))).))))...(((((.........)))))....(....) ( -29.20) >consensus AGUUCCUUAGUCUGUUAUAGCAUUGCAGGGGAUAAGUGAAGGUCUACGAAAUACUAUCCCCUACAAGGCUU____UAAUGACCAAGGCAUAUAAAACGACACAGAUUGAAAUACGCAGA ....((((.(((......(((.(((.(((((((.((((..(.....)....))))))))))).))).))).........))).))))................................ (-19.12 = -19.32 + 0.20)



| Location | 11,103,686 – 11,103,805 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.36 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -26.69 |
| Energy contribution | -27.69 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11103686 119 - 22407834 UCUGAGAAUUUCAAUCUGUGUCGUUUUAUAUGCCUUGGUCAUUAAAUAAAGCCUUGUAGGGGAUAGUAUUUCGUAGAUCCGCACUUAUCCCCUGCAAUGCUAUAACAAACUAAGGAACU ....(((((..((.....))..))))).....(((((((.......((.(((.(((((((((((((.....((......))...))))))))))))).))).))....))))))).... ( -29.80) >DroSec_CAF1 6913 115 - 1 UCUGCGUAUUUCAAUCUGUGUCGUUUUAUAUGCUUUGGUCAUUA----AAGCCUUGUAGGGGAUAGUAUUUCGUAUACCUUCACUUAUCCCCUUCAAUGCUAUAACAGACUAAGGAACU ...((((((........))).)))........((((((((.(((----.(((.(((.((((((((((((.....)))).......)))))))).))).))).)))..)))))))).... ( -29.01) >DroSim_CAF1 7171 115 - 1 UCUGCGUAUUUCAAUCUGUGUCGUUUUAUAUGCUUUGGUCAUUA----AAGCCUUGUAGGGGAUAGUAUUUCGUACACCUUCACUUAUCCCCUGCAAUGCUAUAACAGACUAAGGAACU ...((((((........))).)))........((((((((.(((----.(((.((((((((((((((...............)).)))))))))))).))).)))..)))))))).... ( -32.76) >DroEre_CAF1 7050 115 - 1 CUUGCGUAUUUCAGUCUGUGUCGUUUAAUUUGUCUUGGUCAUUA----AAGCCUUGUAGGGGAUAAUAUUUCGUAGACCUACACCUACCCCCUGCAUUGCUGUAUUAGACUAAGGAAUU ...((((((........))).)))........((((((((..((----.(((..((((((((..........((((........))))))))))))..))).))...)))))))).... ( -29.70) >DroYak_CAF1 6789 115 - 1 CUUGCGUAUUUCAAAUUGUGUCGUUUUGAUUGCCUUGGUCUUUA----AAGCCUUGUAGGGGAUAAUAUUUCGCAGACCUACACCUAUCCCCUGCAUUGCUGUAUUAGACUAAGGAACU ..........(((((.........)))))...(((((((((...----.(((..(((((((((((....................)))))))))))..))).....))))))))).... ( -36.35) >consensus UCUGCGUAUUUCAAUCUGUGUCGUUUUAUAUGCCUUGGUCAUUA____AAGCCUUGUAGGGGAUAGUAUUUCGUAGACCUACACUUAUCCCCUGCAAUGCUAUAACAGACUAAGGAACU ................................((((((((.........(((.(((((((((((((..................))))))))))))).)))......)))))))).... (-26.69 = -27.69 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:45 2006