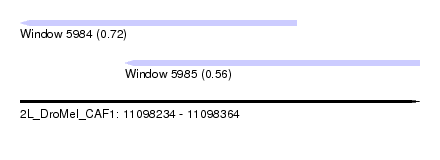
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,098,234 – 11,098,364 |
| Length | 130 |
| Max. P | 0.724014 |

| Location | 11,098,234 – 11,098,324 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.42 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11098234 90 - 22407834 AGCUUCAACGGGCAGUUCAUCUGCCAUGGUGGCCGUGGCGGCAGCCGCGGCUGCGGCACCUUCGGCCAUCCUGCAGGA------CGCCACUAAGGA .(((......(((((.....)))))...(..((((((((....))))))))..)))).((((.(((..(((....)))------.)))...)))). ( -43.60) >DroSec_CAF1 1493 90 - 1 AGCUUCAACGGGCAGUUCAUCUGCCAUAGUGGCCGUGGCGGCAGCCGCGGCUGCUGCACCUUCAGCCAUCCUGCAGGA------CGCCACUAAGGA ..........(((((.....))))).(((((((.((.((((((((....)))))))).(((.(((.....))).))))------)))))))).... ( -42.40) >DroSim_CAF1 1500 90 - 1 AGCUUCAACGGGCAGUUCAUCUGCCAUAGUGGCCGUGGCGGCAGCCGCGGCUGCUGCACCUUCAGCCAUCCUGCAGGA------CGCCACUAAGGA ..........(((((.....))))).(((((((.((.((((((((....)))))))).(((.(((.....))).))))------)))))))).... ( -42.40) >DroEre_CAF1 1491 90 - 1 AGCUUCAACGGGCAACUCGUCUGCCAUAGUGGCCGUGGCGGCAGCUGCAGCGGCUGCACCUUCAGCCAUCCUGCAGGA------CGCCACUAAGGA .(((......((((.......)))).....))).((((((....((((((.(((((......)))))...))))))..------))))))...... ( -36.00) >DroYak_CAF1 1478 90 - 1 AGCUUCAACCGGCAGCUCAUCUGCCAUAGUGGCCGUGGCGGCAGCCGCGGCAGCAGCCCCUUCAGCCAUCCUGCAGGA------CGCCACUAAUGA ....(((...(((((.....))))).(((((((((((((....))))))))....((.(((.(((.....))).))).------.))))))).))) ( -37.10) >DroAna_CAF1 1442 96 - 1 AGCCUCCACCACUAGCUCUUCCGCUGUCGUGGCAGUGGCGGCGGCUGCAGCGGCCGCUCCAUCGGCCAUUCUGCAAGAGGCAGUCGCUAACAAAGA .(((.(..((((((((......))))..))))..).)))(((((((((...(((((......))))).(((.....))))))))))))........ ( -41.00) >consensus AGCUUCAACGGGCAGCUCAUCUGCCAUAGUGGCCGUGGCGGCAGCCGCGGCUGCUGCACCUUCAGCCAUCCUGCAGGA______CGCCACUAAGGA ..........(((((.....))))).(((((((....(((((.((....)).))))).(((.(((.....))).)))........))))))).... (-30.32 = -30.85 + 0.53)

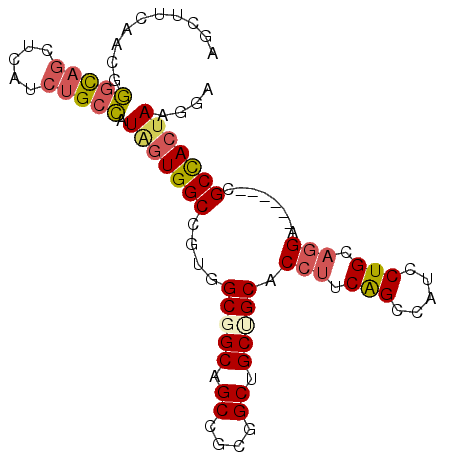
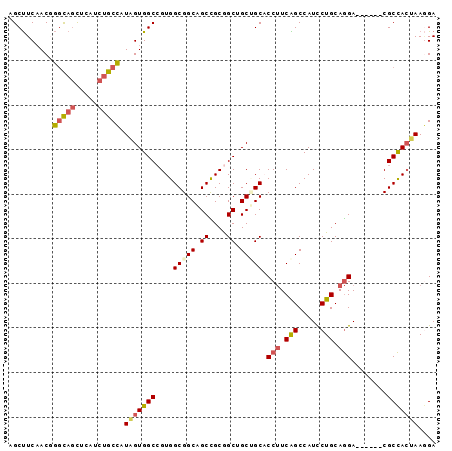
| Location | 11,098,268 – 11,098,364 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.87 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11098268 96 - 22407834 CUGUUCAGCUGGAUGCAUUAUUCGCAAAAUCCGCAAAGAGAGCUUCAACGGGCAGUUCAUCUGCCAUGGUGGCCGUGGC---GGCAGCCGCGGCUGCGG ..((((.((.((((((.......)))...))))).....)))).......(((((.....)))))...(..((((((((---....))))))))..).. ( -38.20) >DroSec_CAF1 1527 96 - 1 CUGUUCAGCUGGAUGCAUUGUUCGCAAAAUCCGCAAAGAGAGCUUCAACGGGCAGUUCAUCUGCCAUAGUGGCCGUGGC---GGCAGCCGCGGCUGCUG ..((((.((.((((...(((....))).)))))).....)))).......(((((.....))))).(((..((((((((---....))))))))..))) ( -40.20) >DroSim_CAF1 1534 96 - 1 CUGUUCAGUUGGAUGCAUUGUUCGCAAAAUCCGCAAAGAGAGCUUCAACGGGCAGUUCAUCUGCCAUAGUGGCCGUGGC---GGCAGCCGCGGCUGCUG .......((((((.((.((.((.((.......)).)).)).)))))))).(((((.....))))).(((..((((((((---....))))))))..))) ( -42.40) >DroEre_CAF1 1525 96 - 1 CUGUACAGCUGGAUGCAUUGUUUGCAAAAUCCGCAAAGAGAGCUUCAACGGGCAACUCGUCUGCCAUAGUGGCCGUGGC---GGCAGCUGCAGCGGCUG ((((.((((((((.((.((.(((((.......))))).)).)).)).((((....).)))(((((((.......)))))---))))))))..))))... ( -32.90) >DroWil_CAF1 1443 93 - 1 ------CACUCGAUGCAUUGUUUGGCAAGUCGGCAAAACGUGCCUCAGCUGGCAGCUCAUCCGCUGUGGUAGCUGUGGCCGUGGCAACUGCAACUGCCG ------.................((((.((..(((..(((.(((.(((((((((((......)))))..)))))).))))))(....)))).)))))). ( -35.00) >DroYak_CAF1 1512 96 - 1 CUGUACAGCUGGAUGCAUUGUUUGCGAAAUCCGCUAAGAGAGCUUCAACCGGCAGCUCAUCUGCCAUAGUGGCCGUGGC---GGCAGCCGCGGCAGCAG ((((...((((((.((.((.((.(((.....))).)).)).)).))....(((((.....))))).)))).((((((((---....)))))))).)))) ( -38.30) >consensus CUGUUCAGCUGGAUGCAUUGUUCGCAAAAUCCGCAAAGAGAGCUUCAACGGGCAGCUCAUCUGCCAUAGUGGCCGUGGC___GGCAGCCGCGGCUGCUG .......((.((((((.......))...))))))................(((((.....))))).(((((((((((((.......))))))))))))) (-26.42 = -26.87 + 0.45)

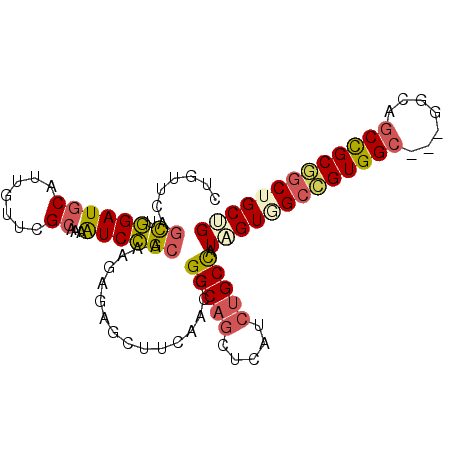
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:35 2006