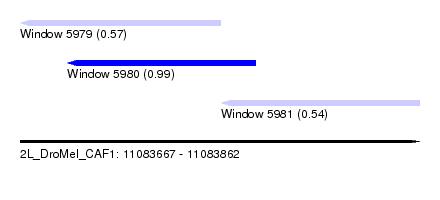
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,083,667 – 11,083,862 |
| Length | 195 |
| Max. P | 0.989997 |

| Location | 11,083,667 – 11,083,765 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 73.43 |
| Mean single sequence MFE | -17.93 |
| Consensus MFE | -11.25 |
| Energy contribution | -12.37 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11083667 98 - 22407834 AAAAGUCGUCUCUGGAGUGGAUUAAAAGCCACAAACAUUUCAGAACCCAACUGAUCGUCAGAGAUGUGUUUUCAAACAAAAAAAAAACCGUUUUGAUA ((((..(((((((((.((((........)))).......((((.......))))...)))))))))..))))....(((((.........)))))... ( -19.80) >DroSec_CAF1 72135 92 - 1 AUAAGCCGUCUCUGGAGUGGACUAAAAACCACAAACAUUUCAAAUCCCAACUGAUCGUUAGAGAUGUGUUUUCAAA------AAUAUAUGUUUUGAUA ..(((((((((((((.((((........)))).......(((.........)))...))))))))).))))(((((------(.......)))))).. ( -18.30) >DroSim_CAF1 73264 92 - 1 AUAAGCCGUCUCUGGAGUGGACUAAAAACCACAAACAUUUCAAAACCCAACUGAUCGUCAGAGAUGUGUUUUCAAA------AAUAAGAGUUUUGAUA ..(((((((((((((.((((........)))).......(((.........)))...))))))))).))))(((((------(.......)))))).. ( -20.40) >DroEre_CAF1 75191 87 - 1 A-AAGCCGUCUCUGGAGUGGACCACAUGCCACAAACCUUUCAAAUCCCAACUGAUCGUCAGAGAUGUGUUUA----------AGAACCCGUUUUGAAA .-(((((((((((((.((((........)))).......(((.........)))...))))))))).)))).----------................ ( -19.40) >DroYak_CAF1 75708 88 - 1 AGAAUCCGUCUCUGGAGUGGACUUAAAGCCACAAACAUUUCAAAUCCCAACUGAUCGUCAGAGAUGUGUUUG----------AAAAACAGUUUUGACA .(((..(((((((((.((((........)))).......(((.........)))...)))))))))..))).----------................ ( -18.60) >DroAna_CAF1 74146 85 - 1 AUAAUUCGCGUCUGGAGUUGGUUUUAAACUAAUAGCAUCCC----CUCAACCCAUCUCCAAUAGUAUUAGUUC---------CGAAACGGUUUUGUUC .......((...(((((.(((....................----......))).)))))...))..(((..(---------((...)))..)))... ( -11.07) >consensus AUAAGCCGUCUCUGGAGUGGACUAAAAACCACAAACAUUUCAAAUCCCAACUGAUCGUCAGAGAUGUGUUUUC_________AAAAACAGUUUUGAUA ..(((((((((((((.((((........)))).......(((.........)))...))))))))).))))........................... (-11.25 = -12.37 + 1.11)


| Location | 11,083,690 – 11,083,782 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -19.58 |
| Consensus MFE | -16.98 |
| Energy contribution | -16.86 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
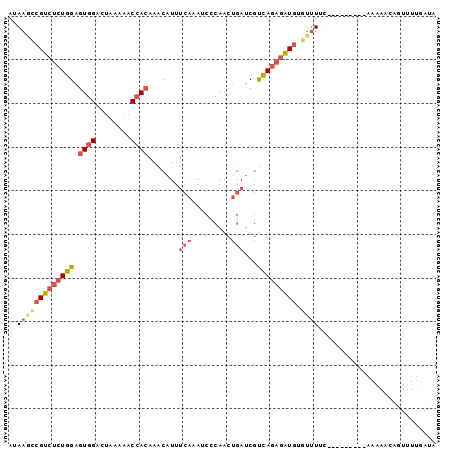
>2L_DroMel_CAF1 11083690 92 - 22407834 AACA---------------------AAAA--AUACAGCAUAAAAGUCGUCUCUGGAGUGGAUUAAAAGCCACAAACAUUUCAGAACCCAACUGAUCGUCAGAGAUGUGUUUUCAA ....---------------------.(((--((((.((......)).((((((((.((((........)))).......((((.......))))...)))))))))))))))... ( -20.50) >DroSec_CAF1 72152 111 - 1 AG--AAACACUGAACAAACACCCAUAAAA--AUACAGCAUAUAAGCCGUCUCUGGAGUGGACUAAAAACCACAAACAUUUCAAAUCCCAACUGAUCGUUAGAGAUGUGUUUUCAA .(--((((((...................--.....((......)).((((((((.((((........)))).......(((.........)))...)))))))))))))))... ( -18.80) >DroSim_CAF1 73281 113 - 1 AAAAAAACACUGAACAAACACCCAUAAAA--AUACAGCAUAUAAGCCGUCUCUGGAGUGGACUAAAAACCACAAACAUUUCAAAACCCAACUGAUCGUCAGAGAUGUGUUUUCAA ...(((((((...................--.....((......)).((((((((.((((........)))).......(((.........)))...)))))))))))))))... ( -20.50) >DroEre_CAF1 75207 96 - 1 ---------------AAACACCCAAAAAAAAGUCCAGCAAA-AAGCCGUCUCUGGAGUGGACCACAUGCCACAAACCUUUCAAAUCCCAACUGAUCGUCAGAGAUGUGUUUA--- ---------------((((((...............((...-..)).((((((((.((((........)))).......(((.........)))...)))))))))))))).--- ( -19.50) >DroYak_CAF1 75724 98 - 1 ---------CUA---AAACACCCAUAAAA--AUCCAGCAAAGAAUCCGUCUCUGGAGUGGACUUAAAGCCACAAACAUUUCAAAUCCCAACUGAUCGUCAGAGAUGUGUUUG--- ---------...---..............--..........(((..(((((((((.((((........)))).......(((.........)))...)))))))))..))).--- ( -18.60) >consensus A________CU____AAACACCCAUAAAA__AUACAGCAUAUAAGCCGUCUCUGGAGUGGACUAAAAGCCACAAACAUUUCAAAUCCCAACUGAUCGUCAGAGAUGUGUUUUCAA ..........................................(((((((((((((.((((........)))).......(((.........)))...))))))))).)))).... (-16.98 = -16.86 + -0.12)


| Location | 11,083,765 – 11,083,862 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -20.89 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
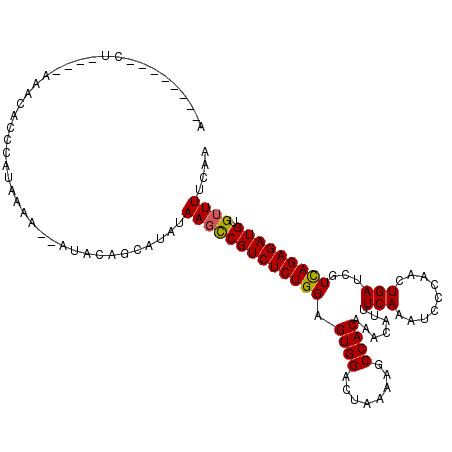
>2L_DroMel_CAF1 11083765 97 - 22407834 GCGUAUCCAACCAUCCGGAUCCGGCGGUGGCCGAGCGUACUGGCGUUGCUAUUUCAAUGCCGUCAGCUGCUUUUAAACAAAACA---------------------AAAA--AUACAGCAU ((((((((........)))).((((....)))).))))...(((((((......)))))))....((((..(((..........---------------------..))--)..)))).. ( -26.10) >DroSec_CAF1 72227 116 - 1 GCGUAUCCAACCAUCCGGAUCCGGCGGUGGCCGAGCCUACUGGCGUUGCUAUUUCAAUGCCGUCAGCUGCUUUUAAACAAAG--AAACACUGAACAAACACCCAUAAAA--AUACAGCAU ((((((........(((....))).((((............(((((((......))))))).((((.((.((((......))--)).)))))).....)))).......--)))).)).. ( -26.70) >DroSim_CAF1 73356 118 - 1 GCGUAUCCAACCAUCCGGAUCCGGCGGUGGCCGAGCCUACUGGCGUUGCUAUUUCAAUGCCGUCAGCUGCUUUUAAACAAAAAAAAACACUGAACAAACACCCAUAAAA--AUACAGCAU ((((((........(((....))).((((............(((((((......))))))).((((.((.((((........)))).)))))).....)))).......--)))).)).. ( -26.40) >DroEre_CAF1 75278 103 - 1 GCGUAUCCAACCAUCCGGAUCCGGCGGUGGACGAGCGUACUGGCGAUGCUAUUUCAAUGCCGUCAGCUGCUUCUGAAA-----------------AAACACCCAAAAAAAAGUCCAGCAA ((.......(((..(((....))).)))(((((((((..((((((..((.........)))))))).))))).((...-----------------...))...........)))).)).. ( -25.60) >DroYak_CAF1 75796 104 - 1 GCGUAUCCAGCCAUCCGGAUCCGGCGGUGGCCGAGCGUACUGGCGUUGCUAUUUCAAUGCCGUCAGCUGCUUCUGAAA-----------CUA---AAACACCCAUAAAA--AUCCAGCAA ((((((((.((((((((....)))..))))).).).)))).(((((((......))))))).((((......))))..-----------...---..............--.....)).. ( -26.60) >DroAna_CAF1 74231 90 - 1 GCGACUCCAUCCAUCUGGAUCCGGCGGUGGUCGUAUGUACUGGCGUUGCUAUUUCAAUGCCGUCAGCUGCUUUUGAAAACCG--AAAAACU----------------------------G .((..((((......))))..)).((((..(((...(((((((((..((.........)))))))).)))...)))..))))--.......----------------------------. ( -25.00) >consensus GCGUAUCCAACCAUCCGGAUCCGGCGGUGGCCGAGCGUACUGGCGUUGCUAUUUCAAUGCCGUCAGCUGCUUUUAAAAAAA________CU____AAACACCCAUAAAA__AUACAGCAU ((....(((.((..(((....))).)))))...(((..((.(((((((......)))))))))..))))).................................................. (-20.89 = -21.12 + 0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:28 2006