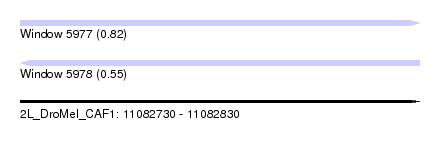
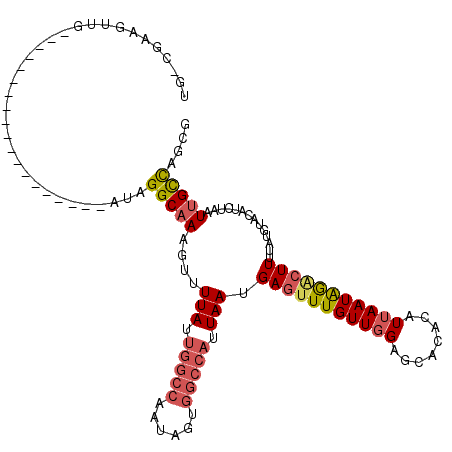
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,082,730 – 11,082,830 |
| Length | 100 |
| Max. P | 0.817115 |

| Location | 11,082,730 – 11,082,830 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -12.73 |
| Energy contribution | -14.03 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11082730 100 + 22407834 CGCUGGCAAUUAGAUGUACAUAAAAGUCUAUUAAUGUGUGCUCCAACAAACUCAUUAAUGGCCACUAUUGGCCAAUAAAACUUUGCCUAU-------------------CAACUUCG-CA ....(((((...((.(((((((............)))))))))...........(((.((((((....)))))).)))....)))))...-------------------........-.. ( -22.00) >DroPse_CAF1 105404 120 + 1 CGAAGGCAAUUAGAUGCACAUAAAAGUUCAUUAAUGUCCGUCCAAACAAUCUCAUUAAUGCGCUCUAUUGGCCAAUAAAACUUUGCCAUUGGCCACCAGAUAAAACUUUCAAAUGCAAAA ..............((((....((((((((((((((................))))))))...(((..((((((((...........))))))))..)))...))))))....))))... ( -23.39) >DroSim_CAF1 72290 100 + 1 CGCUGGCAAUUAGAUGUACAUAAAAGUCUAUUAAUGUGUGCUCCAACAAACUCAUUAAUGGCCACUAUUGGCCAAUAAAACUUUGCCUAU-------------------CAACUUCG-CA ....(((((...((.(((((((............)))))))))...........(((.((((((....)))))).)))....)))))...-------------------........-.. ( -22.00) >DroEre_CAF1 74283 100 + 1 CGCUGGCAAUUAGAUGUACAUAAAAGCCUAUUAAUGUGUGCUCCAACAAACUCAUUAAUGGCCACUAUUGGCCAAUAAAACUUUGCCUAU-------------------CACUUUCG-CA ....(((((...((.(((((((............)))))))))...........(((.((((((....)))))).)))....)))))...-------------------........-.. ( -22.00) >DroYak_CAF1 74829 100 + 1 CGCUGGCAAUUAGCUGUACAUAAAAGUCUAUUAAUGUGUGCUCCAACAAAAUCAUUAAUGGCCACUAUUGGCCAAUAAAACUUUGCCUAU-------------------CAACUUGG-CA .((((((((...(..(((((((............)))))))..)..........(((.((((((....)))))).)))....)))))...-------------------......))-). ( -22.90) >DroAna_CAF1 72994 99 + 1 CGCUAACUAUCUGAUGUACAUAAAAGUCUAUUAAUAGGUGAUCCAACAAACUCAUUAUUGGCCGCUAUA-GCUCAUAAAACUUUGCCAUU-------------------UCACUUGA-CA ...........(((((.((......)).))))).(((((((.....((((....((((.(((.......-))).))))...)))).....-------------------))))))).-.. ( -11.90) >consensus CGCUGGCAAUUAGAUGUACAUAAAAGUCUAUUAAUGUGUGCUCCAACAAACUCAUUAAUGGCCACUAUUGGCCAAUAAAACUUUGCCUAU___________________CAACUUCG_CA ....(((((......(((((((............))))))).............(((.(((((......))))).)))....)))))................................. (-12.73 = -14.03 + 1.31)

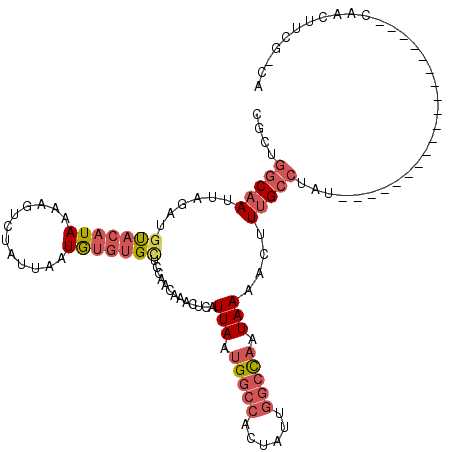
| Location | 11,082,730 – 11,082,830 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -16.28 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11082730 100 - 22407834 UG-CGAAGUUG-------------------AUAGGCAAAGUUUUAUUGGCCAAUAGUGGCCAUUAAUGAGUUUGUUGGAGCACACAUUAAUAGACUUUUAUGUACAUCUAAUUGCCAGCG ..-........-------------------...(((((....(((.((((((....)))))).))).(((((((((((.(....).)))))))))))..............))))).... ( -24.30) >DroPse_CAF1 105404 120 - 1 UUUUGCAUUUGAAAGUUUUAUCUGGUGGCCAAUGGCAAAGUUUUAUUGGCCAAUAGAGCGCAUUAAUGAGAUUGUUUGGACGGACAUUAAUGAACUUUUAUGUGCAUCUAAUUGCCUUCG ...(((((.((((((((...((((.((((((((((.......)))))))))).))))...((((((((...((((....)))).)))))))))))))))).))))).............. ( -35.10) >DroSim_CAF1 72290 100 - 1 UG-CGAAGUUG-------------------AUAGGCAAAGUUUUAUUGGCCAAUAGUGGCCAUUAAUGAGUUUGUUGGAGCACACAUUAAUAGACUUUUAUGUACAUCUAAUUGCCAGCG ..-........-------------------...(((((....(((.((((((....)))))).))).(((((((((((.(....).)))))))))))..............))))).... ( -24.30) >DroEre_CAF1 74283 100 - 1 UG-CGAAAGUG-------------------AUAGGCAAAGUUUUAUUGGCCAAUAGUGGCCAUUAAUGAGUUUGUUGGAGCACACAUUAAUAGGCUUUUAUGUACAUCUAAUUGCCAGCG .(-(....(((-------------------...((((((.(((((.((((((....)))))).))).)).))))))....))).........(((..(((........)))..))).)). ( -28.10) >DroYak_CAF1 74829 100 - 1 UG-CCAAGUUG-------------------AUAGGCAAAGUUUUAUUGGCCAAUAGUGGCCAUUAAUGAUUUUGUUGGAGCACACAUUAAUAGACUUUUAUGUACAGCUAAUUGCCAGCG ..-....((((-------------------...((((((((((((.((((((....)))))).))).)))))))))..(((..((((.((......)).))))...)))......)))). ( -24.60) >DroAna_CAF1 72994 99 - 1 UG-UCAAGUGA-------------------AAUGGCAAAGUUUUAUGAGC-UAUAGCGGCCAAUAAUGAGUUUGUUGGAUCACCUAUUAAUAGACUUUUAUGUACAUCAGAUAGUUAGCG ((-((..(((.-------------------..((((...(((.((.....-)).))).))))((((.(((((((((((........)))))))))))))))...)))..))))....... ( -17.60) >consensus UG_CGAAGUUG___________________AUAGGCAAAGUUUUAUUGGCCAAUAGUGGCCAUUAAUGAGUUUGUUGGAGCACACAUUAAUAGACUUUUAUGUACAUCUAAUUGCCAGCG .................................(((((....(((.(((((......))))).))).(((((((((((........)))))))))))..............))))).... (-16.28 = -17.12 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:25 2006