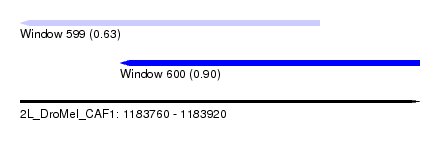
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,183,760 – 1,183,920 |
| Length | 160 |
| Max. P | 0.902641 |

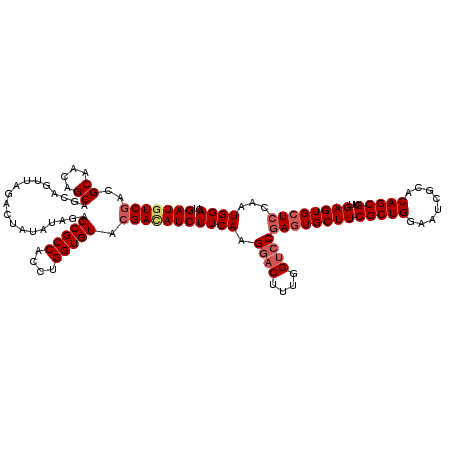
| Location | 1,183,760 – 1,183,880 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -41.18 |
| Consensus MFE | -36.70 |
| Energy contribution | -37.46 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1183760 120 - 22407834 CACCUGGUGUACGACAUCUUCAAGAACUUUGGUCCGAGUGCUUCGCUGGAAUCGCACAGCGUUGAGUGCUCCAAUGGACUGGACGGCUUCAUGUCCGCCUUGUACACCGUUACGCUGGAC .....(((((((((................((((((((..((((((((........)))))..)))..)))....)))))(((((......)))))...)))))))))............ ( -42.80) >DroSec_CAF1 17156 120 - 1 CAUCUGGUGUACGACAUCUUCAAGGACUUUGGUCCGAGUGCUUCGCUGGAAUCGCACAGCGUUGAGUGCUCCAAUGGACUGGACGGCUUCAAGUAUACCUUGUACAUAGUUACGAAGGAC ..((((((((((....(((....))).((..(((((((..((((((((........)))))..)))..)))....))))..)).........)))))))(((((......))))).))). ( -37.30) >DroSim_CAF1 17107 120 - 1 CACCUGGUGUACGACAUCUUCAAGGACUUUGGUCCGAGUGCUUCGCUGGAAUCGCACAGCGUUGAGUGCUCCAAUGGACUGGAUGGCUUCAUGUCCGCCUUGUACACCGUUACGCUGGAC .....(((((((((.........((((....))))(((..((((((((........)))))..)))..)))....((((((((....)))).))))...)))))))))............ ( -43.70) >DroEre_CAF1 18225 120 - 1 CACCUGGUGUACGAUAUCUUCAAGGACUUUGGACCGAGUGCUUCGCUGGAAUCGCACAGCGUAGAGUGCUCCAAUGGACUGGACGGCUUCAUGUCCGCCCUGUACACCGUUACGCUGGAC .....(((((((((.....))..((((..(((((((.(..((((((((........)))))..)))..)((((......))))))).)))).)))).....)))))))............ ( -42.00) >DroYak_CAF1 17884 120 - 1 CACCUGGUGUACGAAAUCUUCAAGGACUUCGGUCCCAAUGCUUCGCUGGAAUCGCACAGCGUAGAGUGCUCCAAUGGACUGGACGGCUUCAUGUCCGCCUUGUACACCAUUACGCUGGAC ....((((((((((.........((((....))))....((((((.((((...((((........)))))))).))))..(((((......))))))).))))))))))........... ( -40.10) >consensus CACCUGGUGUACGACAUCUUCAAGGACUUUGGUCCGAGUGCUUCGCUGGAAUCGCACAGCGUUGAGUGCUCCAAUGGACUGGACGGCUUCAUGUCCGCCUUGUACACCGUUACGCUGGAC ....((((((((((.........((((....))))(((((((((((((........)))))..))))))))...(((((((((....)))).)))))..))))))))))........... (-36.70 = -37.46 + 0.76)

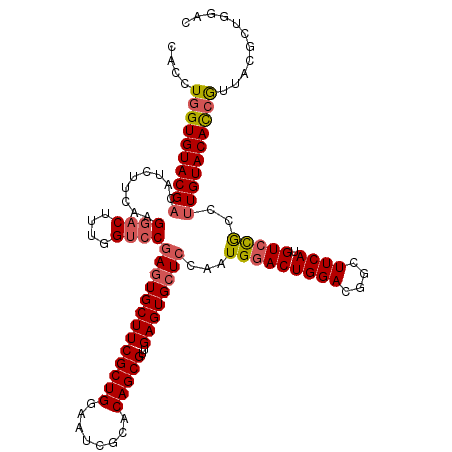
| Location | 1,183,800 – 1,183,920 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -36.12 |
| Energy contribution | -37.20 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1183800 120 - 22407834 GAUGUCGACGCAACAGCAGCAGUUAGACUAUAUAGAGCGCCACCUGGUGUACGACAUCUUCAAGAACUUUGGUCCGAGUGCUUCGCUGGAAUCGCACAGCGUUGAGUGCUCCAAUGGACU (((((((..((....))...................(((((....))))).)))))))............((((((((..((((((((........)))))..)))..)))....))))) ( -40.90) >DroSec_CAF1 17196 120 - 1 GAUGUCGACGCAACAGCAGCAGUUAGACUAUAUAGAGCGCCAUCUGGUGUACGACAUCUUCAAGGACUUUGGUCCGAGUGCUUCGCUGGAAUCGCACAGCGUUGAGUGCUCCAAUGGACU (((((((..((....))...................(((((....))))).)))))))((((.((((....))))(((..((((((((........)))))..)))..)))...)))).. ( -42.90) >DroSim_CAF1 17147 120 - 1 GAUGUCGACGCAACAGCAGCAGUUAGACUAUAUAGAGCGCCACCUGGUGUACGACAUCUUCAAGGACUUUGGUCCGAGUGCUUCGCUGGAAUCGCACAGCGUUGAGUGCUCCAAUGGACU (((((((..((....))...................(((((....))))).)))))))((((.((((....))))(((..((((((((........)))))..)))..)))...)))).. ( -42.90) >DroEre_CAF1 18265 120 - 1 GAUGUCGACGCAACAGCAGCAGCUGGACUAUAUCGAGCGCCACCUGGUGUACGAUAUCUUCAAGGACUUUGGACCGAGUGCUUCGCUGGAAUCGCACAGCGUAGAGUGCUCCAAUGGACU (((((((((((..(((..((.(((.((.....)).)))))...))))))).)))))))....((..(.(((((....(..((((((((........)))))..)))..)))))).)..)) ( -40.90) >DroYak_CAF1 17924 120 - 1 GAUGUCAACGCAACAGCAGCAGUUAGACUAUAUCGAGCGCCACCUGGUGUACGAAAUCUUCAAGGACUUCGGUCCCAAUGCUUCGCUGGAAUCGCACAGCGUAGAGUGCUCCAAUGGACU ...(((.......((((((((....((..((.(((.(((((....))))).))).))..))..((((....))))...))))..)))).....((((........)))).......))). ( -36.90) >consensus GAUGUCGACGCAACAGCAGCAGUUAGACUAUAUAGAGCGCCACCUGGUGUACGACAUCUUCAAGGACUUUGGUCCGAGUGCUUCGCUGGAAUCGCACAGCGUUGAGUGCUCCAAUGGACU (((((((..((....))...................(((((....))))).)))))))((((.((((....))))(((((((((((((........)))))..))))))))...)))).. (-36.12 = -37.20 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:27 2006