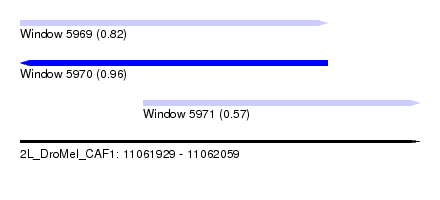
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,061,929 – 11,062,059 |
| Length | 130 |
| Max. P | 0.955624 |

| Location | 11,061,929 – 11,062,029 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
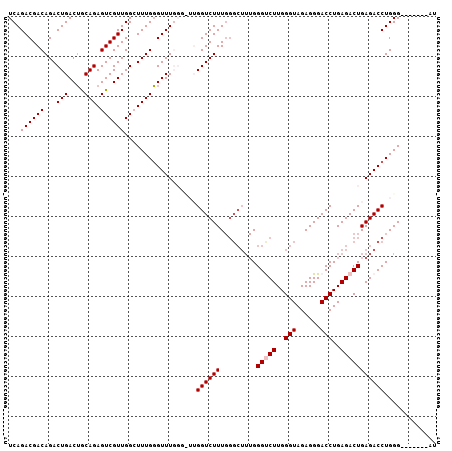
>2L_DroMel_CAF1 11061929 100 + 22407834 UCAGACGACAGACUGACUGCAGAGUCGUUGGCUUUGGGUUUGGGCUUGGUCUUUGGGCUUUGGGCCUUGGGUAGAGGGACCUGAGACUGAGACCUGGG-------AU ...((((((...(((....))).))))))...((..(((((.((((((((((((((((.....)))).......))))))).))).)).)))))..))-------.. ( -33.61) >DroSec_CAF1 50621 94 + 1 UCAGACGACAGACUGACUGCAGAGUCGUUGGCUUUGGGUUU------GGUCUUUGGGCUUUGGGUCUUGGGUAGAGGGACCUGAGACUGAGACCUGGG-------AU .........((((..(.(.((((((.....)))))).).).------.))))(..((.(((.(((((..(((......)))..))))).)))))..).-------.. ( -35.10) >DroSim_CAF1 50838 94 + 1 UCAGACGACAGACUGACUGCAGAGUCGUUGGCUUUGGGUUU------GGUCUUUGGGCUUUGGGUCUUGGGUAGAGGGACCUGAGACUGAGACCUGGG-------AU .........((((..(.(.((((((.....)))))).).).------.))))(..((.(((.(((((..(((......)))..))))).)))))..).-------.. ( -35.10) >DroEre_CAF1 53367 107 + 1 UCAGACGACAGACUGACUGCAGAGUCGUUGGGUUUGGGUUUGGGUUUGGUCUUUGGGCUUUGGGCCUUGGGUAGAGGGACCUGAGACUGAGACCUGGGAUCUGGGAU (((((((((.((((........))))))))..((..(((((.((((((((((((((((.....)))).......)))))))..))))).)))))..)).)))))... ( -36.91) >DroYak_CAF1 53067 93 + 1 UCAGACGACAGACUGACAACAGAGUCGUUGGGUUUGUGUUUGGGUUUGGUCUUU-------GGGUCUUGGGUAGAGGGACCUGAGACUGAGACCUGGG-------AU ((((((.(((((((..((((......)))))))))))))))))....((((((.-------.(((((..(((......)))..)))))))))))....-------.. ( -36.50) >consensus UCAGACGACAGACUGACUGCAGAGUCGUUGGCUUUGGGUUUGGG_UUGGUCUUUGGGCUUUGGGUCUUGGGUAGAGGGACCUGAGACUGAGACCUGGG_______AU ...((((((...(((....))).))))))..................((((((.........(((((..(((......)))..)))))))))))............. (-25.62 = -26.02 + 0.40)


| Location | 11,061,929 – 11,062,029 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -19.79 |
| Consensus MFE | -17.84 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
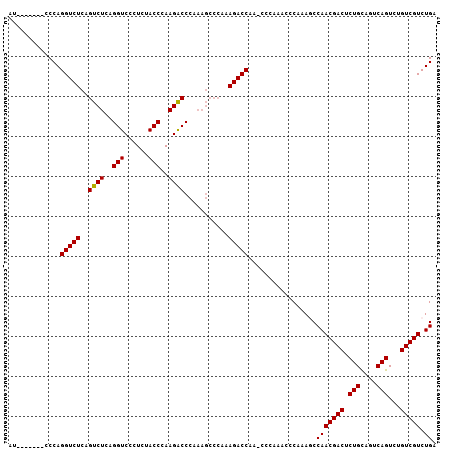
>2L_DroMel_CAF1 11061929 100 - 22407834 AU-------CCCAGGUCUCAGUCUCAGGUCCCUCUACCCAAGGCCCAAAGCCCAAAGACCAAGCCCAAACCCAAAGCCAACGACUCUGCAGUCAGUCUGUCGUCUGA ..-------....((.((..((((..(((......)))...(((.....)))...))))..)).))...........(((((((.(((....)))...))))).)). ( -19.20) >DroSec_CAF1 50621 94 - 1 AU-------CCCAGGUCUCAGUCUCAGGUCCCUCUACCCAAGACCCAAAGCCCAAAGACC------AAACCCAAAGCCAACGACUCUGCAGUCAGUCUGUCGUCUGA ..-------....(((((..((((..(((......)))..))))...........)))))------...........(((((((.(((....)))...))))).)). ( -18.32) >DroSim_CAF1 50838 94 - 1 AU-------CCCAGGUCUCAGUCUCAGGUCCCUCUACCCAAGACCCAAAGCCCAAAGACC------AAACCCAAAGCCAACGACUCUGCAGUCAGUCUGUCGUCUGA ..-------....(((((..((((..(((......)))..))))...........)))))------...........(((((((.(((....)))...))))).)). ( -18.32) >DroEre_CAF1 53367 107 - 1 AUCCCAGAUCCCAGGUCUCAGUCUCAGGUCCCUCUACCCAAGGCCCAAAGCCCAAAGACCAAACCCAAACCCAAACCCAACGACUCUGCAGUCAGUCUGUCGUCUGA ....(((((..(((((((........(((......)))...(((.....)))...))))).....................((((....))))....))..))))). ( -21.00) >DroYak_CAF1 53067 93 - 1 AU-------CCCAGGUCUCAGUCUCAGGUCCCUCUACCCAAGACCC-------AAAGACCAAACCCAAACACAAACCCAACGACUCUGUUGUCAGUCUGUCGUCUGA ..-------....(((((..((((..(((......)))..))))..-------..))))).................(((((((.(((....)))...))))).)). ( -22.10) >consensus AU_______CCCAGGUCUCAGUCUCAGGUCCCUCUACCCAAGACCCAAAGCCCAAAGACCAA_CCCAAACCCAAAGCCAACGACUCUGCAGUCAGUCUGUCGUCUGA .............(((((..((((..(((......)))..))))...........))))).................(((((((.(((....)))...))))).)). (-17.84 = -17.60 + -0.24)

| Location | 11,061,969 – 11,062,059 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -27.60 |
| Energy contribution | -27.93 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
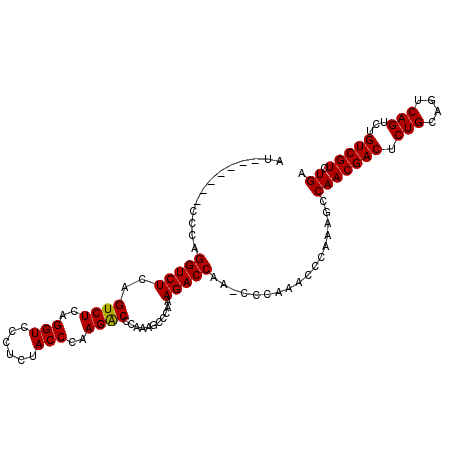
>2L_DroMel_CAF1 11061969 90 + 22407834 UGGGCUUGGUCUUUGGGCUUUGGGCCUUGGGUAGAGGGACCUGAGACUGAGACCUGGGAUCUGAGACCAAGUCAGGCCAAGUCAAGUCGA ..((((((((((((((..((..(..((..(((...((.........))...)))..))..)..)).))))...))))))))))....... ( -34.60) >DroSec_CAF1 50661 84 + 1 U------GGUCUUUGGGCUUUGGGUCUUGGGUAGAGGGACCUGAGACUGAGACCUGGGAUCUGAGACCAAGUCAGGCCAAGUCAAGUCGA (------((((((..((.(((.(((((..(((......)))..))))).)))))..)((..((....))..))))))))........... ( -30.00) >DroSim_CAF1 50878 84 + 1 U------GGUCUUUGGGCUUUGGGUCUUGGGUAGAGGGACCUGAGACUGAGACCUGGGAUCUGAGACCAAGUCAGGCCAAGUCAACUCGA (------((((((..((.(((.(((((..(((......)))..))))).)))))..)((..((....))..))))))))........... ( -30.00) >consensus U______GGUCUUUGGGCUUUGGGUCUUGGGUAGAGGGACCUGAGACUGAGACCUGGGAUCUGAGACCAAGUCAGGCCAAGUCAAGUCGA .......(((((((((..((..(((((..(((...((.........))...)))..)))))..)).))))...)))))............ (-27.60 = -27.93 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:17 2006