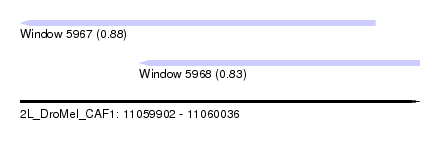
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,059,902 – 11,060,036 |
| Length | 134 |
| Max. P | 0.883457 |

| Location | 11,059,902 – 11,060,021 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.76 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11059902 119 - 22407834 UUACAAGUGCAAUAAAAAGUGGUUGACAAGUAGUUGGGCAAAUAGUGCA-UCUGAAAUGUGUCAAAAACAACAGUGGGCUAACACUUGUGUCUGAACUGAAAUACAUAUCAAGUCGGGUU .((((((((..........((.(..((.....))..).))..((((.((-.(((...(((.......))).))))).)))).))))))))(((((..(((........)))..))))).. ( -27.90) >DroSec_CAF1 48600 120 - 1 UUACAAGUGCAAUAAAAAGUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAAAAGUGGGCUAACACUUGUGUCUGAACUGAAAUACAUAUCAAGUCGGGUU .((((((((.........((..((((((..((((((.((.....)).))).))).....))))))..))...((....))..))))))))(((((..(((........)))..))))).. ( -28.20) >DroSim_CAF1 48750 120 - 1 UUACAAGUGCAAUAAAAAGUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAACAGUGGGCUAACACUUGUGUCUGAACUGAAAUACAUAUCAAGUCGGCUU .((((((((.........((..((((((..((((((.((.....)).))).))).....))))))..))...((....))..)))))))).((((..(((........)))..))))... ( -26.90) >DroEre_CAF1 51074 120 - 1 UUACAAGUGCAAUAAAGUGUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAACAGUGGGCUAACACUUGUGUCUAGGCUGAAAAACAUAUCAAGUCGGGUU .((((((((........(((..((((((..((((((.((.....)).))).))).....))))))..)))..((....))..))))))))....((((((........)).))))..... ( -26.80) >DroYak_CAF1 50766 119 - 1 UUACAAGUGCAAUAAAA-GUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAACAGUGGGCUAACACUUGUGCCUAAGCUGAAAAACAUAUCAAGUCGGGUU ..(((((((........-((..((((((..((((((.((.....)).))).))).....))))))..))...((....))..)))))))(((..(..(((........)))..)..))). ( -25.80) >consensus UUACAAGUGCAAUAAAAAGUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAACAGUGGGCUAACACUUGUGUCUGAACUGAAAUACAUAUCAAGUCGGGUU .((((((((.........((..((((((..((((((.((.....)).))).))).....))))))..))...((....))..))))))))(((((..(((........)))..))))).. (-25.24 = -25.76 + 0.52)


| Location | 11,059,942 – 11,060,036 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 97.16 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11059942 94 - 22407834 UGAUUUAUUGCUGCUUUACAAGUGCAAUAAAAAGUGGUUGACAAGUAGUUGGGCAAAUAGUGCA-UCUGAAAUGUGUCAAAAACAACAGUGGGCU ...((((((((.(((.....)))))))))))..((..((((((..(((.((.((.....)).))-.))).....))))))..))........... ( -22.10) >DroSec_CAF1 48640 95 - 1 UGAUUUACUGCUGCUUUACAAGUGCAAUAAAAAGUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAAAAGUGGGCU .......(..((((.........))........((..((((((..((((((.((.....)).))).))).....))))))..))...))..)... ( -19.20) >DroSim_CAF1 48790 95 - 1 UGAUUUAUUGCUGCUUUACAAGUGCAAUAAAAAGUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAACAGUGGGCU ...((((((((.(((.....)))))))))))..((..((((((..((((((.((.....)).))).))).....))))))..))........... ( -22.40) >DroEre_CAF1 51114 95 - 1 UGAUUUAUUGCUGCUUUACAAGUGCAAUAAAGUGUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAACAGUGGGCU ...((((((((.(((.....))))))))))).(((..((((((..((((((.((.....)).))).))).....))))))..))).......... ( -23.10) >DroYak_CAF1 50806 94 - 1 UGAUUUAUCGCUGCUUUACAAGUGCAAUAAAA-GUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAACAGUGGGCU .......(((((((.........)).......-((..((((((..((((((.((.....)).))).))).....))))))..))...)))))... ( -19.00) >consensus UGAUUUAUUGCUGCUUUACAAGUGCAAUAAAAAGUGGUUGACAAGUAGUUGGGCAAAUAGUGCAAUCUGAAAUGUGUCAAAAACAACAGUGGGCU ...((((((((.(((.....)))))))))))..((..((((((..((((((.((.....)).))).))).....))))))..))........... (-20.10 = -20.70 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:14 2006