| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,051,630 – 11,051,732 |
| Length | 102 |
| Max. P | 0.842161 |

| Location | 11,051,630 – 11,051,732 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -14.63 |
| Energy contribution | -14.43 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
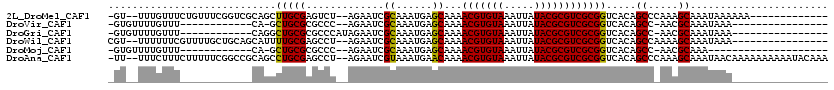
>2L_DroMel_CAF1 11051630 102 + 22407834 -------------UUUUUUAUUUGCUUUGGGCUGUGACCGCGACGCGUAUAAUUUACACGUUUUGCUCAUUUGCGAUUCU--AGACUCGCAAGCUGCGACCGAAACAGAAACAAA--AC- -------------.......((((.(((((((.((((.((.((((.(((.....))).)))).)).))))..))......--....((((.....))))))))).))))......--..- ( -25.00) >DroVir_CAF1 56487 87 + 1 ----------------UUUAUUUGCGUU-GGCUGUGACCGCGACGCGUAUAAUUUACACGUUUUGCUCAUUUGCGAUUCU--GGGCGCGCAGC-UG------------AAACAAAACAC- ----------------....((((..(.-.((((((..(((((((.(((.....))).))))((((......))))....--..)))))))))-..------------)..))))....- ( -27.20) >DroGri_CAF1 50043 90 + 1 ----------------UUUAUUUGCGUU-GGCUGUGACCGCGACGCGUAUAAUUUACACGUUUUGCUCAUUUGCGAUUCUAUGGGCGCGCAGCCUG------------AAACAAAACAC- ----------------....((((..((-(((((((..(((((((.(((.....))).))))((((......))))........)))))))))).)------------)..))))....- ( -25.90) >DroWil_CAF1 37343 100 + 1 ----------------UUUAUUUGCUUUUGGCUGUGACCGCGACGCGUAUAAUUUACACGUUUUGCUCAUUUGCGAUUCU--AGGCUCGCAAAAUGCUGCAGCAAAACGAAAAAA--ACG ----------------....((((.(((((.((((((.((.((((.(((.....))).)))).)).)).(((((((....--....))))))).....)))))))))))))....--... ( -24.90) >DroMoj_CAF1 52126 83 + 1 --------------------UUUGCGUU-GGCUGUGACCGCGACGCGUAUAAUUUACACGUUUUGCUCAUUUGCGAUUCU--GGGCGCGCAGC-UG------------AAACAAAACAC- --------------------((((..(.-.((((((..(((((((.(((.....))).))))((((......))))....--..)))))))))-..------------)..))))....- ( -26.90) >DroAna_CAF1 45965 115 + 1 UUUGUAUUUUUUUUUUGUUAUUUGCUUUGGGCUGUGACCGCGACGCGUAUAAUUUACACGUUUUGUUCAUUUACGAUUCU--AGGCUCGCAGGCUGCGGCCGAAAAAGAAAGAAA--AA- ......((((((((((((.....))(((.((((((..((((((.(((((.....)))..(..((((......))))..).--..)))))).))..)))))).)))))))))))))--..- ( -31.00) >consensus ________________UUUAUUUGCGUU_GGCUGUGACCGCGACGCGUAUAAUUUACACGUUUUGCUCAUUUGCGAUUCU__AGGCGCGCAGCCUG____________AAACAAA__AC_ .......................((.....)).((((.((.((((.(((.....))).)))).)).))))(((((..((....))..)))))............................ (-14.63 = -14.43 + -0.19)


| Location | 11,051,630 – 11,051,732 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.65 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.03 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.842161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
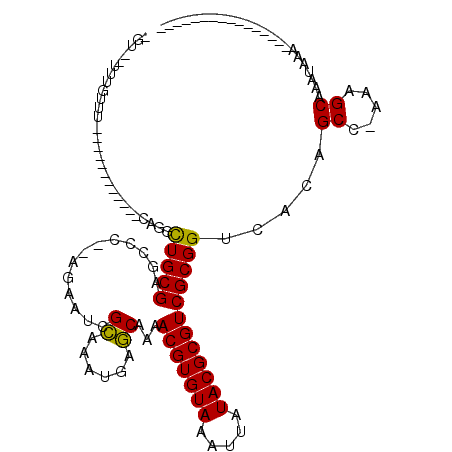
>2L_DroMel_CAF1 11051630 102 - 22407834 -GU--UUUGUUUCUGUUUCGGUCGCAGCUUGCGAGUCU--AGAAUCGCAAAUGAGCAAAACGUGUAAAUUAUACGCGUCGCGGUCACAGCCCAAAGCAAAUAAAAAA------------- -((--((((...((((...(..(((.((((((((....--....)))))....)))...(((((((.....))))))).)))..)))))..))))))..........------------- ( -31.60) >DroVir_CAF1 56487 87 - 1 -GUGUUUUGUUU------------CA-GCUGCGCGCCC--AGAAUCGCAAAUGAGCAAAACGUGUAAAUUAUACGCGUCGCGGUCACAGCC-AACGCAAAUAAA---------------- -(((((..(((.------------..-(((((((((..--......((......)).....(((((...)))))))).))))))...))).-))))).......---------------- ( -22.30) >DroGri_CAF1 50043 90 - 1 -GUGUUUUGUUU------------CAGGCUGCGCGCCCAUAGAAUCGCAAAUGAGCAAAACGUGUAAAUUAUACGCGUCGCGGUCACAGCC-AACGCAAAUAAA---------------- -(((((..(((.------------..((((((((((..........((......)).....(((((...)))))))).)))))))..))).-))))).......---------------- ( -24.20) >DroWil_CAF1 37343 100 - 1 CGU--UUUUUUCGUUUUGCUGCAGCAUUUUGCGAGCCU--AGAAUCGCAAAUGAGCAAAACGUGUAAAUUAUACGCGUCGCGGUCACAGCCAAAAGCAAAUAAA---------------- .((--((((...(((.((((((.((..(((((((....--....)))))))...))...(((((((.....))))))).)))).)).))).)))))).......---------------- ( -29.00) >DroMoj_CAF1 52126 83 - 1 -GUGUUUUGUUU------------CA-GCUGCGCGCCC--AGAAUCGCAAAUGAGCAAAACGUGUAAAUUAUACGCGUCGCGGUCACAGCC-AACGCAAA-------------------- -(((((..(((.------------..-(((((((((..--......((......)).....(((((...)))))))).))))))...))).-)))))...-------------------- ( -22.30) >DroAna_CAF1 45965 115 - 1 -UU--UUUCUUUCUUUUUCGGCCGCAGCCUGCGAGCCU--AGAAUCGUAAAUGAACAAAACGUGUAAAUUAUACGCGUCGCGGUCACAGCCCAAAGCAAAUAACAAAAAAAAAAUACAAA -..--..............((((((....(((((....--....)))))..........(((((((.....))))))).))))))...((.....))....................... ( -24.00) >consensus _GU__UUUGUUU____________CAGGCUGCGAGCCC__AGAAUCGCAAAUGAGCAAAACGUGUAAAUUAUACGCGUCGCGGUCACAGCC_AAAGCAAAUAAA________________ ............................(((((.............((......))...(((((((.....)))))))))))).....((.....))....................... (-14.53 = -14.03 + -0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:05 2006