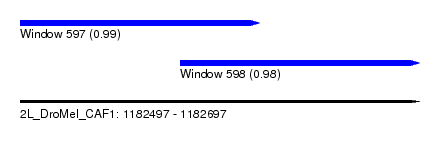
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,182,497 – 1,182,697 |
| Length | 200 |
| Max. P | 0.989384 |

| Location | 1,182,497 – 1,182,617 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -45.52 |
| Consensus MFE | -42.16 |
| Energy contribution | -43.08 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1182497 120 + 22407834 AAUCUCGUCCAUGUAGCCAUGCUCAUAGGCGUGACGAACGGUCUUGAAGAUCUCCUGGUUCGCCUCAUCGCCAGCGAUGUCCAAGGACAACUGGUGGAAGGCGGGGCCGUGCAUAUCCGU ..((.((((.(((.(((...)))))).)))).)).(.(((((((....((((....))))(((((..(((((((...((((....)))).))))))).)))))))))))).)........ ( -43.20) >DroSec_CAF1 15890 120 + 1 AAUCUCGUCCAUGUAGCCAUGCUCAUAGGCGUGGCGCACGGUCUUGAAGAUCUCCUGGUUCGCCUCAUCGCCUGCGAUGUCCAAGGACAACUGGUGGAAGGCGGGGCCGUGCAUAUCCGU ...............((((((((....))))))))(((((((((((....((..(..(((..((((((((....)))))....)))..)))..)..))...)))))))))))........ ( -50.40) >DroSim_CAF1 15842 120 + 1 AAUCUCGUCCAUGUAGCCAUGCUCAUAGGCGUGGCGCACGAUCUUGAAGAUCUCCUGGUUCGCCUCAUCGCCUGCGAUGUCCAAGGACAACUGGUGGAAAGCGGGGCCGUGCAUAUCCGU ...............((((((((....))))))))(((((..((((....((..(..(((..((((((((....)))))....)))..)))..)..))...))))..)))))........ ( -44.70) >DroEre_CAF1 16951 120 + 1 AAUCUCGUCCAUGUAGCCAUGCUCAUAGGCAUGGCGCACGGUCUUGAAAAUCUCCUGGUUCGCCUCAUCGCCUCCGAUGUCCAAGGACAACUGGUGGAAGGCAGGGCCGUGCAUAUCCGU ...............((((((((....))))))))(((((((((((....((..(..(((..((((((((....)))))....)))..)))..)..))...)))))))))))........ ( -49.90) >DroYak_CAF1 16610 120 + 1 AAUCGCGUCCAUGUAGCCAUGCUCAUAGGCAUGCCGCACGGUCUUGAAAAUCUCCUGGUUCGCCACAUCGCCUCCAAUGUCCAAGGACAACUGGUGGAAGGCAGGCCCGUGCAUAUCGGU .((((((((((((.(((...)))))).)).)))).((((((.(((..........(((....)))....(((((((.((((....)))).....))).))))))).)))))).....))) ( -39.40) >consensus AAUCUCGUCCAUGUAGCCAUGCUCAUAGGCGUGGCGCACGGUCUUGAAGAUCUCCUGGUUCGCCUCAUCGCCUGCGAUGUCCAAGGACAACUGGUGGAAGGCGGGGCCGUGCAUAUCCGU ...............((((((((....))))))))(((((((((((....((..(..(((..((((((((....)))))....)))..)))..)..))...)))))))))))........ (-42.16 = -43.08 + 0.92)


| Location | 1,182,577 – 1,182,697 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -48.74 |
| Consensus MFE | -41.80 |
| Energy contribution | -42.40 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1182577 120 + 22407834 CCAAGGACAACUGGUGGAAGGCGGGGCCGUGCAUAUCCGUCUCGAAGAGGCGGGACAACUCGGCGCAAUCCUUCUCGGUGCCGAGCAGCCAGGGAAGGAAGUGCAUGCAUACCAUUGGUC .........((..((((..(((...)))((((((.((((((((...))))))))........((((..(((((((((((((...))).)).)))))))).)))))))))).))))..)). ( -50.00) >DroSec_CAF1 15970 120 + 1 CCAAGGACAACUGGUGGAAGGCGGGGCCGUGCAUAUCCGUCUCGAAGAGGCGGGACAACUCGGCGCAAUCCUUCUCGGUGCCAAGCAGCCAGGGAAGGAAGUGCAUGCCUACCAUUGGUC .........((..((((.(((((..((((.(....((((((((...))))))))....).))))(((.(((((((((((((...))).)).))))))))..))).))))).))))..)). ( -54.80) >DroSim_CAF1 15922 120 + 1 CCAAGGACAACUGGUGGAAAGCGGGGCCGUGCAUAUCCGUCUCGAAGAGGCGGGACAGCUCGGCGCAAUCCUUCUCGGUGCCAAGCAGCCAGGGAAGGAAGUGCAUGCAUACCAUUGGUC .........((..((((...(((..((((.((...((((((((...))))))))...)).))))(((.(((((((((((((...))).)).))))))))..))).)))...))))..)). ( -52.20) >DroEre_CAF1 17031 120 + 1 CCAAGGACAACUGGUGGAAGGCAGGGCCGUGCAUAUCCGUUUCGAAGAGGCGGGACAACUCGGCGCAAUCCUUCUCAGCACCGAGCAGCCAGGGCAGGAAGUGCAUGCACACCAUUGGUC .....(((...(((((...(((...)))((((((.(((((..((......))..))..(((((.((...........)).)))))..((....)).))).))))))...)))))...))) ( -43.00) >DroYak_CAF1 16690 120 + 1 CCAAGGACAACUGGUGGAAGGCAGGCCCGUGCAUAUCGGUUUCGAAGAGGCGGGACAACUCGGCGCAAUCCUUCUCAGAGCUGAGCAGCCAGGGAAGGAAGUGCAUGCAUACCAUUGGUC .........((..((((...(((.((.(((((......((..((......))..))......))))..((((((((.(.(((....)))).)))))))).).)).)))...))))..)). ( -43.70) >consensus CCAAGGACAACUGGUGGAAGGCGGGGCCGUGCAUAUCCGUCUCGAAGAGGCGGGACAACUCGGCGCAAUCCUUCUCGGUGCCGAGCAGCCAGGGAAGGAAGUGCAUGCAUACCAUUGGUC .........((..((((...(((..((((.(....((((((((...))))))))....).))))(((.(((((((((((........))).))))))))..))).)))...))))..)). (-41.80 = -42.40 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:24 2006