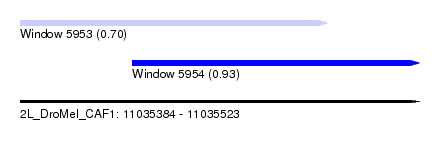
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,035,384 – 11,035,523 |
| Length | 139 |
| Max. P | 0.926465 |

| Location | 11,035,384 – 11,035,491 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.37 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.62 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
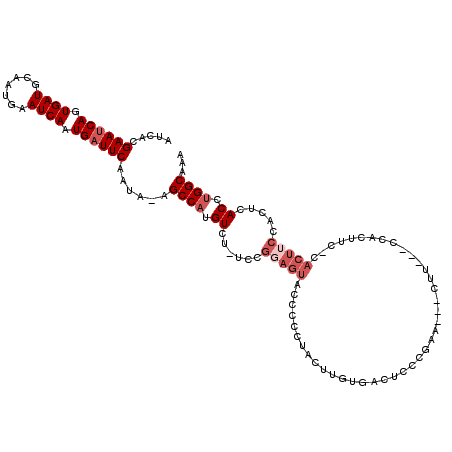
>2L_DroMel_CAF1 11035384 107 + 22407834 UAAUGAAUCAU-UCGAACAGAUCACAUGGGUCACGGCUAUAUCACGAAUCAGUGAUGCAAUGAAUCAAUGAUUCAAUAAAGCCAUGUCUCUCCGGAGUGU------------ACCCCCGA ...........-...............(((((((((((.(((...((((((.((((.......)))).)))))).))).))))...(((....)))))).------------)))).... ( -25.70) >DroSec_CAF1 23855 119 + 1 UAAUGAAUCAUUUCGCACAGAUCACAUGGGUCACGGUUAUAUCACGAAUCAGUGAUGCAAUGAAUCAAUGAUUCAAUA-AGCCAUGUCUGUCCGUAGUACCCCCUACUUGUGACUCCCGA ....(((....))).............(((((((((((.((((((......))))))...((((((...))))))...-))))..........((((......))))..))))))).... ( -27.60) >DroSim_CAF1 23884 119 + 1 UAAUGAAUCAUUUCGAACAGAUCACAUGGGUCACGGUUAUAUCACGAAUCAGUGAUGCAAUGAAUCAAUGAUUCAAUA-AGCCAUGUCUGUCCGUAGUACCCCCUACUUGUGACUCCCGA ...((((....))))............(((((((((((.((((((......))))))...((((((...))))))...-))))..........((((......))))..))))))).... ( -27.90) >DroEre_CAF1 25544 115 + 1 UAAUGAAUCACUUCAAACUAAUCACAUGGGUCACGGCUAUAUCACGAACCAGUGAUGCAAUAAAUCAAUGAUUCAAUA-AGCCCAGUCU----GGAGUACCCCCUAGUUGUGGCUCCCAA ...(((((((..........(((((.(((....((.........))..))))))))............)))))))...-(((((((.((----((........))))))).))))..... ( -25.65) >DroYak_CAF1 24729 114 + 1 -AAUGAAUCAUUUUCACCUGAUUACAUGGGUCACGGUUAUAUUACGAAUCAGUGAUGCAAUAAAUCAAUGAUUCAAUA-AGCCAUGUCU----GGAGUACCACCUAGUUGUGACUCCCAA -.((((((((........))))).)))(((((((((((.((((..((((((.((((.......)))).))))))))))-))))....((----((........))))..))))))).... ( -29.50) >consensus UAAUGAAUCAUUUCGAACAGAUCACAUGGGUCACGGUUAUAUCACGAAUCAGUGAUGCAAUGAAUCAAUGAUUCAAUA_AGCCAUGUCU_UCCGGAGUACCCCCUACUUGUGACUCCCGA ...........................(((((((((((.(((...((((((.((((.......)))).)))))).))).))))..(((((....))).)).........))))))).... (-21.46 = -21.62 + 0.16)



| Location | 11,035,423 – 11,035,523 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -12.52 |
| Energy contribution | -14.12 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11035423 100 + 22407834 AUCACGAAUCAGUGAUGCAAUGAAUCAAUGAUUCAAUAAAGCCAUGUCUCUCCGGAGUGU------------ACCCCCGAA---CUU---CCACUUC-CACUUCCACUCACCUGGCAAA .....((((((.((((.......)))).))))))......((((.((......((((((.------------.........---...---.......-)))))).....)).))))... ( -21.25) >DroSec_CAF1 23895 111 + 1 AUCACGAAUCAGUGAUGCAAUGAAUCAAUGAUUCAAUA-AGCCAUGUCUGUCCGUAGUACCCCCUACUUGUGACUCCCGAA---CUU---CCACUUC-CACUUCGACUCACCUGGCAAA .....((((((.((((.......)))).))))))....-.((((.((..(((.((((......))))..(((.........---...---.)))...-......)))..)).))))... ( -24.92) >DroSim_CAF1 23924 105 + 1 AUCACGAAUCAGUGAUGCAAUGAAUCAAUGAUUCAAUA-AGCCAUGUCUGUCCGUAGUACCCCCUACUUGUGACUCCCGAA---CUU---CCACUUC-CA------CUCACCUGGCAAA .....((((((.((((.......)))).))))))....-.((((.((..((..((((......))))..(((.........---...---.)))...-.)------)..)).))))... ( -21.32) >DroEre_CAF1 25584 102 + 1 AUCACGAACCAGUGAUGCAAUAAAUCAAUGAUUCAAUA-AGCCCAGUCU----GGAGUACCCCCUAGUUGUGGCUCCCAAA---CCC---------CAAACUUCCACUCACCUGGCAAA .....(((.((.((((.......)))).)).)))....-(((((((.((----((........))))))).))))......---...---------.......(((......))).... ( -17.10) >DroYak_CAF1 24768 114 + 1 AUUACGAAUCAGUGAUGCAAUAAAUCAAUGAUUCAAUA-AGCCAUGUCU----GGAGUACCACCUAGUUGUGACUCCCAAGACACCCCUCGCCCUUCACCCUUCCACUCACCUGGCAAA .....((((((.((((.......)))).))))))....-.(((((((((----(((((..(((......))))))))..)))))............................))))... ( -25.88) >consensus AUCACGAAUCAGUGAUGCAAUGAAUCAAUGAUUCAAUA_AGCCAUGUCU_UCCGGAGUACCCCCUACUUGUGACUCCCGAA___CUU___CCACUUC_CACUUCCACUCACCUGGCAAA .....((((((.((((.......)))).))))))......((((.((......(((((.........................................))))).....)).))))... (-12.52 = -14.12 + 1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:57 2006