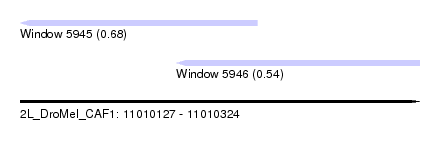
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,010,127 – 11,010,324 |
| Length | 197 |
| Max. P | 0.683075 |

| Location | 11,010,127 – 11,010,244 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.77 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -29.98 |
| Energy contribution | -31.54 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11010127 117 - 22407834 CCAGAAGGCGGGCAAUCACCAGGGCUUCUGCAUUUGGUUCGAUGUACAGUUUCCUGGCGAAGAU---UUUGUUCUAAGCACAUCGCCGCUAUCCCCGCCAACGCAUUGGAAGCAGUGUGU ......((((((.....((((((((....)).)))))).((((((.(((....)))((..(((.---.....)))..))))))))........)))))).((((((((....)))))))) ( -42.10) >DroSec_CAF1 5006 117 - 1 CCAGAAGGCGGGCAAUCACCAGGGCUUCUGCAUUUGGUUCGAUGUACGGUUUCCUGGCGAAGAA---UUUGUUCUAAGCACAUCGCCGCUCUCCCCGCCAACGCACUGGAAGCAGUGUGU ......((((((...((.((((((..((((((((......)))))).))..)))))).))((((---....)))).(((........)))...)))))).((((((((....)))))))) ( -44.00) >DroSim_CAF1 5528 117 - 1 CCAGAAGGCGGGCAAUCACCAGGGCUUCUGCAUUUGGUUCGAUGUACGGUUUCCUGGCGAAGAA---UUUGUUCUGAGCACAUCGCCGCUCUCCCCGCCAACGCAUUGGAAGCAGUGUGU ......((((((...((.((((((..((((((((......)))))).))..)))))).))((((---....))))((((........))))..)))))).((((((((....)))))))) ( -45.40) >DroYak_CAF1 5001 117 - 1 GCAGAAGGCGGGCAACCAUCAGGGCUUCUGCAUUUGGUUUGAUGUACGAUUCCCUGGCGAAGAU---UUUGUUCUAAGCACAUCCCCACUUUCCACGCCAACGCAUUGGAAGCAGUGUGU ((.(((((.(((.......(((((..((((((((......)))))).))..)))))((..(((.---.....)))..)).....))).)))))...))..((((((((....)))))))) ( -39.80) >DroPer_CAF1 5222 120 - 1 ACAAAGAGGGGGAAACCAUCAAGGAUUUUGCAUAUGGUUUGAGGUGCUAUUUCCAGGCAAUGAAGCCGUGAUUCUUAGCACAUCUCCUUUCGCCCCGGAAACGCAUUGGAAGCAGUGUGU .......((((((((((.....))...........((...((.((((((..((..(((......)))..))....)))))).)).)))))).))))....((((((((....)))))))) ( -37.70) >consensus CCAGAAGGCGGGCAAUCACCAGGGCUUCUGCAUUUGGUUCGAUGUACGGUUUCCUGGCGAAGAA___UUUGUUCUAAGCACAUCGCCGCUCUCCCCGCCAACGCAUUGGAAGCAGUGUGU ......((((((.....((((((((....)).))))))..(((((.(((....)))((..((((.......))))..))))))).........)))))).((((((((....)))))))) (-29.98 = -31.54 + 1.56)

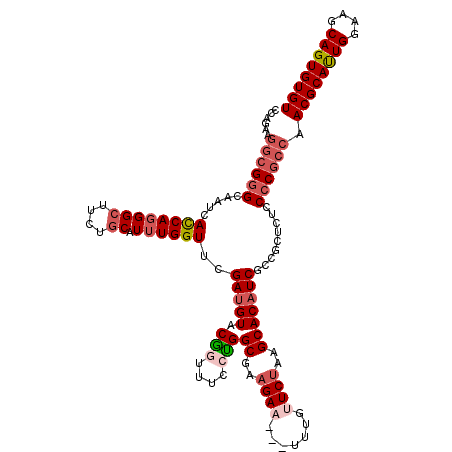

| Location | 11,010,204 – 11,010,324 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -40.36 |
| Consensus MFE | -31.94 |
| Energy contribution | -31.62 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11010204 120 - 22407834 GUCGUCUUUCACUGGAUGAAUCUGCUGGAUGUGGAAGCCAGCGAUUUGGACAGCAUUCAGUUCAAGGAGGUAAUAACAGCCCAGAAGGCGGGCAAUCACCAGGGCUUCUGCAUUUGGUUC .((((((......))))))....((..((((..((((((..(..(((((((........)))))))..).........((((.......)))).........))))))..))))..)).. ( -43.20) >DroSec_CAF1 5083 120 - 1 GUCGUCUUUCACUGGAUGAAUCUGCUGGAUGUGGAGGCCAGCGAUUUGGACAGCAUUCAGUUCAAGGAGGUUAUAACAGCCCAGAAGGCGGGCAAUCACCAGGGCUUCUGCAUUUGGUUC .((((((......))))))....((..((((..((((((......((((((........))))))((.((((......((((.......)))))))).))..))))))..))))..)).. ( -42.90) >DroSim_CAF1 5605 120 - 1 GUCGUCUUUCACAGGAUGAAUCUGCUGGAUGUGGAGGCCAGCGAUUUGGACAGCAUUCAGUUCAAGGAGGUUAUAACAGCCCAGAAGGCGGGCAAUCACCAGGGCUUCUGCAUUUGGUUC .((((((......))))))....((..((((..((((((......((((((........))))))((.((((......((((.......)))))))).))..))))))..))))..)).. ( -42.70) >DroYak_CAF1 5078 120 - 1 GUCGUCUUUCACUGGAUGAAUCUGCUGGAUGUGGAGGCCAGCGAUUUGGACAGCAUUCAGUUUAAGGAGGUGAUAACAGCGCAGAAGGCGGGCAACCAUCAGGGCUUCUGCAUUUGGUUU .((((((......))))))....((..((((..((((((.(((.(((((((........)))))))...((....))..)))....(..((....))..)..))))))..))))..)).. ( -41.40) >DroPer_CAF1 5302 120 - 1 GUCGUUCUUCAUUGGAUGAACUUGCUCGAUGUGGAUAUGGCUGAACUUGAGGAAGUUCGUUUUAAGGAUGUCGUUGCGGCACAAAGAGGGGGAAACCAUCAAGGAUUUUGCAUAUGGUUU (.(((((((....((((((((((.(((((.((............))))))).)))))))))).))))))).)..(((((..(...((..((....)).))...)...)))))........ ( -31.60) >consensus GUCGUCUUUCACUGGAUGAAUCUGCUGGAUGUGGAGGCCAGCGAUUUGGACAGCAUUCAGUUCAAGGAGGUUAUAACAGCCCAGAAGGCGGGCAAUCACCAGGGCUUCUGCAUUUGGUUC .((((((......))))))....((..((((..((((((.((..(((((((........)))))))..((((.....))))......))((....)).....))))))..))))..)).. (-31.94 = -31.62 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:48 2006