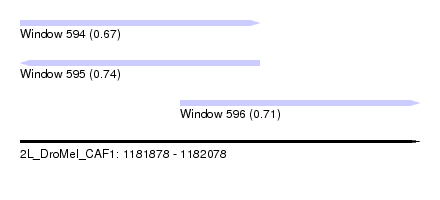
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,181,878 – 1,182,078 |
| Length | 200 |
| Max. P | 0.736610 |

| Location | 1,181,878 – 1,181,998 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -25.43 |
| Energy contribution | -26.72 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1181878 120 + 22407834 CAGGCUGAAGUACGGUGAGAGCGAUCCUCCUCCACCAAAUAGGAGCCGGGAGCGUUUCGAGCAAGAGAUAAAGACUUUGCAGUCGCGCCAGAAGGAUUCUUUUGGCGCCACUGCAGCCAU ..(((((((((..((((.(((.(.....)))))))).....((((((....).)))))...............)))))(((((.((((((((((....)))))))))).))))))))).. ( -50.00) >DroSim_CAF1 15225 120 + 1 CAGGCUGAAGUACGGUGAGAGCGAUCCUCCUCCACCAAACAGGAGCCGGGAGCGUUUCGAGCAGGAGAUAAAGACUUUGCAGUCGCGCCAGAAGGAUUCUUUUGGCGCUACUCCAGCCAC ..((((...((.(((((.(((.(.....)))))))).....((((((....).)))))).)).((((.....((((....))))((((((((((....))))))))))..)))))))).. ( -48.70) >DroEre_CAF1 16332 120 + 1 CAGGCUGAAGUACGGAGAAAGCGAUCCUCCUCCCCCAAAUAAGAGUCGGGAGCGUUUCGAGCAGGAGAUAAAGACUUUGCAGUCGCGCCAGAAGGAUUCUUCUGGGGCUACUCCAGCCAU ..((((...((.(((((...((..(((..(((..........)))..))).)).))))).)).((((.....((((....))))((.(((((((....))))))).))..)))))))).. ( -44.60) >DroYak_CAF1 15995 120 + 1 CAGGCUGAAGUACGGAGAGAGCGAUCCGCCUCCACCGAAUAAAAGCCGGGAGCGUUUCGAGCAGGAGAUAAAGACUUUGCAGUCGCGCCAGAAGGAUUCUUCUGGCGCUACUCCAGCCAU ..(((((.((((((((........)))).((((.(((.........)))..((.......)).)))).....((((....))))((((((((((....)))))))))))))).))))).. ( -49.50) >DroAna_CAF1 15361 111 + 1 GAGAUUGAAGUAUGGGGAGAGUGAUCCUCCGCCACCAAACCGCAGCCGAGAGCACUUCGAGCAGCAGAUGAAGACUCUCCAGUC-CUCCA--------CAUCUGCGUCCACUCCAUCCGU (((.(((((((.(((((((.......)))).))).......((........)))))))))...(((((((..((((....))))-.....--------))))))).....)))....... ( -31.60) >DroPer_CAF1 20075 120 + 1 GAGACUGAAGUACGGGGAGAGCGAUCCACCGCCUCCGAAUCGCAGUCGGGAGCGUUUCGAGCAGGAGAUAAAGACCUUGCAGACGCGUCAGAAGAAUAAUGCCAGCGCGGCUUCAGCUCU (((.(((((((....((((.(((......)))))))....(((.((.((.(((((((...(((((..........))))))))))).((....))....).)).))))))))))))))). ( -41.90) >consensus CAGGCUGAAGUACGGAGAGAGCGAUCCUCCUCCACCAAAUAGGAGCCGGGAGCGUUUCGAGCAGGAGAUAAAGACUUUGCAGUCGCGCCAGAAGGAUUCUUCUGGCGCUACUCCAGCCAU ..(((((.(((.......(((((.(((.....................))).)))))...(((((..........)))))....((((((((((....)))))))))).))).))))).. (-25.43 = -26.72 + 1.28)
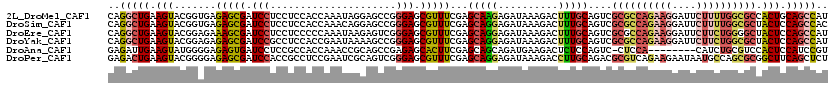
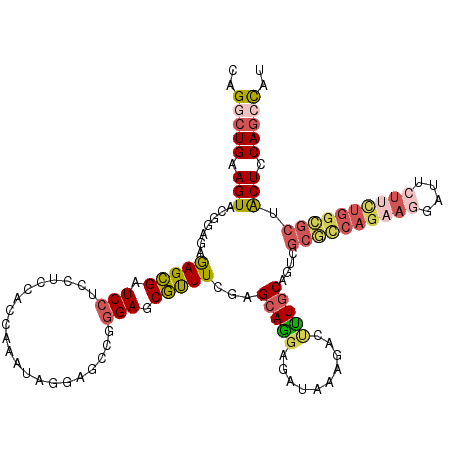
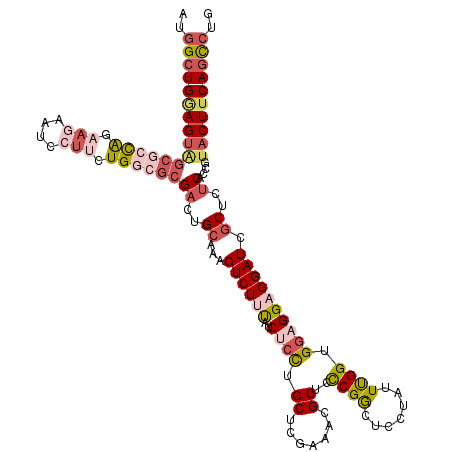
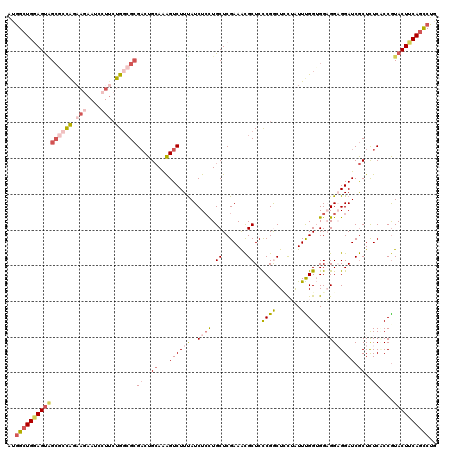
| Location | 1,181,878 – 1,181,998 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -45.65 |
| Consensus MFE | -26.92 |
| Energy contribution | -29.12 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1181878 120 - 22407834 AUGGCUGCAGUGGCGCCAAAAGAAUCCUUCUGGCGCGACUGCAAAGUCUUUAUCUCUUGCUCGAAACGCUCCCGGCUCCUAUUUGGUGGAGGAGGAUCGCUCUCACCGUACUUCAGCCUG ..(((((((((.((((((.(((....))).)))))).)))))..))))..........((.......))....((((......(((((((((.....).))).)))))......)))).. ( -42.60) >DroSim_CAF1 15225 120 - 1 GUGGCUGGAGUAGCGCCAAAAGAAUCCUUCUGGCGCGACUGCAAAGUCUUUAUCUCCUGCUCGAAACGCUCCCGGCUCCUGUUUGGUGGAGGAGGAUCGCUCUCACCGUACUUCAGCCUG ..((((((((((((((((.(((....))).))))))((((....))))....((((((.(.(.((((((.....))....)))).).).)))))).............)))))))))).. ( -48.00) >DroEre_CAF1 16332 120 - 1 AUGGCUGGAGUAGCCCCAGAAGAAUCCUUCUGGCGCGACUGCAAAGUCUUUAUCUCCUGCUCGAAACGCUCCCGACUCUUAUUUGGGGGAGGAGGAUCGCUUUCUCCGUACUUCAGCCUG ..((((((((((((.(((((((....))))))).))((((....))))....................(((((((.......))))))).((((((.....)))))).)))))))))).. ( -49.70) >DroYak_CAF1 15995 120 - 1 AUGGCUGGAGUAGCGCCAGAAGAAUCCUUCUGGCGCGACUGCAAAGUCUUUAUCUCCUGCUCGAAACGCUCCCGGCUUUUAUUCGGUGGAGGCGGAUCGCUCUCUCCGUACUUCAGCCUG ..((((((((((((((((((((....))))))))))((((....)))).............(((..(((..(((.(........).)))..)))..))).........)))))))))).. ( -54.00) >DroAna_CAF1 15361 111 - 1 ACGGAUGGAGUGGACGCAGAUG--------UGGAG-GACUGGAGAGUCUUCAUCUGCUGCUCGAAGUGCUCUCGGCUGCGGUUUGGUGGCGGAGGAUCACUCUCCCCAUACUUCAAUCUC ..((((((((((..(((....)--------))..(-(...(((((((..((.(((((..(.((((.(((........))).)))))..))))).))..))))))))).)))))).)))). ( -44.30) >DroPer_CAF1 20075 120 - 1 AGAGCUGAAGCCGCGCUGGCAUUAUUCUUCUGACGCGUCUGCAAGGUCUUUAUCUCCUGCUCGAAACGCUCCCGACUGCGAUUCGGAGGCGGUGGAUCGCUCUCCCCGUACUUCAGUCUC .(((((((((.((((..((..........))..))))...(((.((.........)))))(((((.(((........))).)))))..((((.(((......))))))).)))))).))) ( -35.30) >consensus AUGGCUGGAGUAGCGCCAGAAGAAUCCUUCUGGCGCGACUGCAAAGUCUUUAUCUCCUGCUCGAAACGCUCCCGGCUCCUAUUUGGUGGAGGAGGAUCGCUCUCACCGUACUUCAGCCUG ..((((((((((((((((.(((....))).))))))((..((...((((((..((((.((.......))..((((.......)))).)))))))))).))..))....)))))))))).. (-26.92 = -29.12 + 2.20)

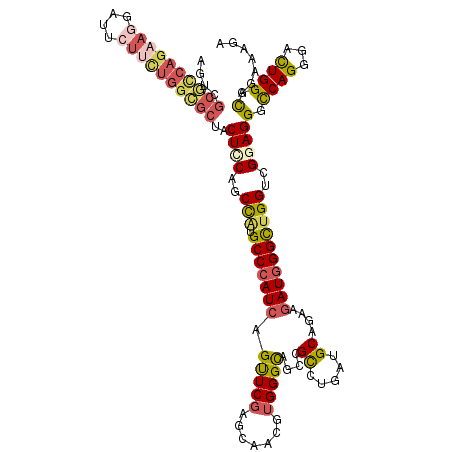
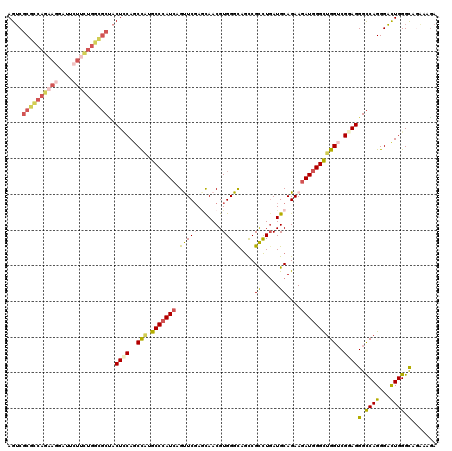
| Location | 1,181,958 – 1,182,078 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -47.85 |
| Consensus MFE | -37.44 |
| Energy contribution | -38.37 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1181958 120 + 22407834 AGUCGCGCCAGAAGGAUUCUUUUGGCGCCACUGCAGCCAUGCCCAUCAGUUCGACCAACGUGGGCAGCCGCCUGAUGCAGAAGAUGGGCUGGUCGGAGGGCCAGGGACUGGGCAAAAAGA ((((.(((((((((....))))))))(((.((.(.((((.(((((((..(((...((...(((((....))))).))..)))))))))))))).).)))))..).))))........... ( -50.60) >DroSim_CAF1 15305 120 + 1 AGUCGCGCCAGAAGGAUUCUUUUGGCGCUACUCCAGCCACGCCCAUCAGUUCGAGCAACGUGGGCAGCCGCCUGAUGCAGAAGAUGGGCUGGUCGCAGGGCCAGGGACUGGGCAAAAAAA (((.((((((((((....)))))))))).)))((((((..(((((((..(((..(((...(((((....))))).))).))))))))))(((((....))))).)).))))......... ( -55.60) >DroEre_CAF1 16412 120 + 1 AGUCGCGCCAGAAGGAUUCUUCUGGGGCUACUCCAGCCAUGCCCAUCAGUUCGAGCAACGUGGGCAGUCGCCUCAUGCAGAAGAUGGGUUGGUCGGAGGGCCAGGGAUUGGGCAGGAAGA ((((.(.(((((((....)))))))((((.((((.((((.(((((((..(((..(((....((((....))))..))).)))))))))))))).)))))))).).))))........... ( -52.70) >DroYak_CAF1 16075 120 + 1 AGUCGCGCCAGAAGGAUUCUUCUGGCGCUACUCCAGCCAUGCCCAUCAGUUCGAGCAACGUGGGCAGUCGCCUGAUGCAGAAGAUGGGUUGGUCGGAGGGCCAGGGAUUGGGCAGGAAGA ....((((((((((....))))))))))..((((.((((.(((((((..(((..(((...(((((....))))).))).)))))))))))))).)))).(((........)))....... ( -57.20) >DroAna_CAF1 15441 111 + 1 AGUC-CUCCA--------CAUCUGCGUCCACUCCAUCCGUGCCCAUCAGCUCUAGCAACGUGGGUAGCCGUCUGAUGCAGAAAAUGGGCUGGUCGGAGGGUCAGGGAUUGGGUAGAAAGA ....-.....--------..(((((..(((.(((.....(((((((..((....))...)))))))(((.((((((.(((........))))))))).)))...))).)))))))).... ( -36.90) >DroPer_CAF1 20155 120 + 1 AGACGCGUCAGAAGAAUAAUGCCAGCGCGGCUUCAGCUCUGCCAAUCAGCUCGAAUAACGUGGGCAAUCGCUUGAUGCAGAAGAUGGGCUGGACGGAGGGCCAGGGUCUGGGCAGGAAGA ...(((((..(..........)..))))).((((..((((((((..(.(((((.......)))))....((.....))........)..))).)))))(.((((...)))).)..)))). ( -34.10) >consensus AGUCGCGCCAGAAGGAUUCUUCUGGCGCUACUCCAGCCAUGCCCAUCAGUUCGAGCAACGUGGGCAGCCGCCUGAUGCAGAAGAUGGGCUGGUCGGAGGGCCAGGGACUGGGCAGAAAGA ....((((((((((....))))))))))..((((..(((.(((((((.(((((.......)))))....((.....))....))))))))))..))))(.((((...)))).)....... (-37.44 = -38.37 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:22 2006