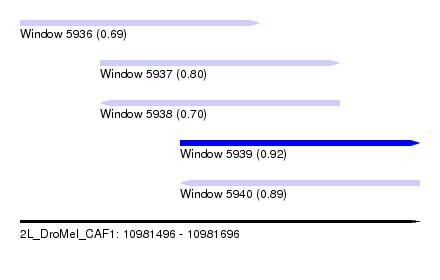
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,981,496 – 10,981,696 |
| Length | 200 |
| Max. P | 0.923925 |

| Location | 10,981,496 – 10,981,616 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -24.95 |
| Energy contribution | -24.07 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
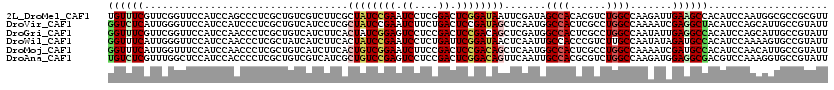
>2L_DroMel_CAF1 10981496 120 + 22407834 ACGUAAAGUUGAAAAGUCAGAGCAAAAGGAAAUUGAACGAAACGCGGCGCCAUUGGAUGUGGCUUCAAUCUUGGCCAGACGUGUGGCUAUCGAAUUAUCCGAGUCCGAGGAUUCGGAUAG .((...(((((....(((.(.((...((((..(((((((.....))..(((((.....)))))))))))))).))).)))...)))))..))...((((((((((....)))))))))). ( -35.50) >DroVir_CAF1 7354 120 + 1 ACGCAAGGUAGAAAAGUCCGAGCUGAAGGAAAUUGAACGCAAUACGGCAAUGCUGGAUGUAGCCUCGAUUUUGGCCAGGCGAGUGGCCAUUGAGCUAUCGGAGUCAGAAGAUUCGGAUAG .(....)........((((.((((.((.((((((((..((.(((((((...))))..))).)).))))))))(((((......))))).)).))))....(((((....))))))))).. ( -34.40) >DroGri_CAF1 8415 120 + 1 GCGAAAGGUGGAAAAGUCGGAGCUAAAGGAAAUUGAACGUAAUACGGCAAUGCUGGAUGUGGCCUCAAUAUUGGCCAGGCGAGUGGCCAUCGAGCUGUCGGAGUCGGAGGACUCCGAUAG ((....(((((...(.(((.................((((....((((...)))).))))((((........))))...))).)..)))))..))((((((((((....)))))))))). ( -42.40) >DroWil_CAF1 9327 120 + 1 GCGAAAGGUGGAGAAAUCCGAGCAAAAGGAAAUUGAACGUAAUACGGCACUUUUGGAUGUGGCAUCUAUAUUGGCAAGACGGGUGGCAAUUGAGUUAUCCGAAUCAGAGGAUUCGGAUAG ((......((((....)))).)).......(((((..(((......((.....((((((...)))))).....))...))).....)))))....((((((((((....)))))))))). ( -31.30) >DroMoj_CAF1 7648 120 + 1 GCGGAAGGUAGAAAAGUCGGAGUUGAAGGAAAUUGAACGCAAUACGGCAAUGUUGGAUGUGGCAUCGAUUUUGGCCAGGCGAGUGGCCAUUGAGCUGUCGGAGUCGGAAGAUUCCGACAG ((....(((...((((((((.(((.((.....)).)))((.((.((((...)))).))...)).)))))))).))).(((.....))).....))((((((((((....)))))))))). ( -42.30) >DroAna_CAF1 9325 120 + 1 GCGAAAGGUGGAAAAGUCUGAGCAGAAGGAAUUUGAACGUAAUACGGCACCUUUGGACGUCGCCUCCAUCUUGGCCAGACGCGUGGCAAUUGAACUGUCCGAGUCGGAGGACUCGGACAG ((.(((((((......(((....)))...........((.....)).))))))).(.(((((((........)))..)))))...)).......(((((((((((....))))))))))) ( -41.00) >consensus GCGAAAGGUGGAAAAGUCCGAGCAAAAGGAAAUUGAACGUAAUACGGCAAUGUUGGAUGUGGCCUCAAUAUUGGCCAGACGAGUGGCCAUUGAGCUAUCCGAGUCGGAGGAUUCGGAUAG ...................((((..............((.....))(((........))).)).)).....((((((......)))))).....(((((((((((....))))))))))) (-24.95 = -24.07 + -0.88)

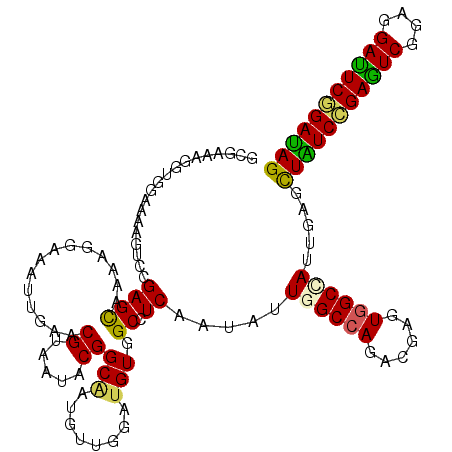
| Location | 10,981,536 – 10,981,656 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -29.80 |
| Energy contribution | -28.80 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10981536 120 + 22407834 AACGCGGCGCCAUUGGAUGUGGCUUCAAUCUUGGCCAGACGUGUGGCUAUCGAAUUAUCCGAGUCCGAGGAUUCGGAUAGCGAAGACGACAGCGAGGGCUGGAUGGAACCGAACGAAACA ....(((.(((((.....)))))(((.((((.((((...(((((.(((.(((...((((((((((....)))))))))).))))).).))).))..)))))))).))))))......... ( -43.60) >DroVir_CAF1 7394 120 + 1 AAUACGGCAAUGCUGGAUGUAGCCUCGAUUUUGGCCAGGCGAGUGGCCAUUGAGCUAUCGGAGUCAGAAGAUUCGGAUAGCGAGGAUGACAGCGAGGGAUGGAUGGAACCCAAUGAGACC .....(((..(((((....((.((((.....((((((......))))))....((((((.(((((....))))).)))))))))).)).))))).(((..........))).....).)) ( -42.10) >DroGri_CAF1 8455 120 + 1 AAUACGGCAAUGCUGGAUGUGGCCUCAAUAUUGGCCAGGCGAGUGGCCAUCGAGCUGUCGGAGUCGGAGGACUCCGAUAGUGAAGAUGACAGCGAGGGUUGGAUGGAACCGAACGAAACC ....(((....(((.(.(.(((((........))))).)).)))..(((((..((((((((((((....))))))))))))........((((....)))))))))..)))......... ( -48.20) >DroWil_CAF1 9367 120 + 1 AAUACGGCACUUUUGGAUGUGGCAUCUAUAUUGGCAAGACGGGUGGCAAUUGAGUUAUCCGAAUCAGAGGAUUCGGAUAGUGAAGAUGAUAGCGAGGGUUGGAUGGAACCCAAUGAAACC ......((.....((((((...)))))).....))......(((..((......(((((((((((....)))))))))))...............(((((......)))))..))..))) ( -31.50) >DroMoj_CAF1 7688 120 + 1 AAUACGGCAAUGUUGGAUGUGGCAUCGAUUUUGGCCAGGCGAGUGGCCAUUGAGCUGUCGGAGUCGGAAGAUUCCGACAGUGAAGAUGACAGCGAGGGUUGGAUGGAAACCAAUGAAACC .....((...((((((......((((.....((((((......))))))....((((((((((((....))))))))))))...)))).((((....))))........))))))...)) ( -45.70) >DroAna_CAF1 9365 120 + 1 AAUACGGCACCUUUGGACGUCGCCUCCAUCUUGGCCAGACGCGUGGCAAUUGAACUGUCCGAGUCGGAGGACUCGGACAGCGAUGACGACAGCGAGGGGUGGAUGGAGCCAAACGAGACA .....(((......((......))(((((((..(((...(((((.(..((((..(((((((((((....)))))))))))))))..).)).)))...))))))))))))).......... ( -48.70) >consensus AAUACGGCAAUGUUGGAUGUGGCCUCAAUAUUGGCCAGACGAGUGGCCAUUGAGCUAUCCGAGUCGGAGGAUUCGGAUAGCGAAGAUGACAGCGAGGGUUGGAUGGAACCCAACGAAACC ..((((.((....))..)))).((((.....((((((......))))))((..((((((((((((....))))))))))))...)).......))))....................... (-29.80 = -28.80 + -1.00)

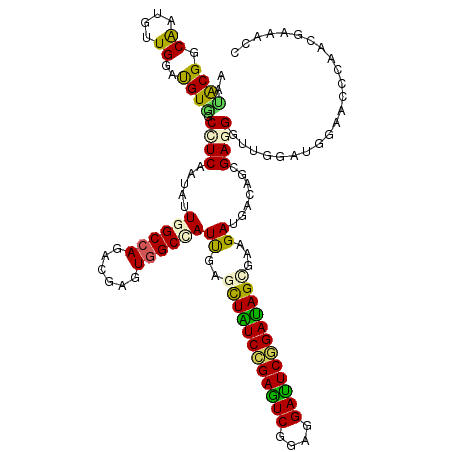
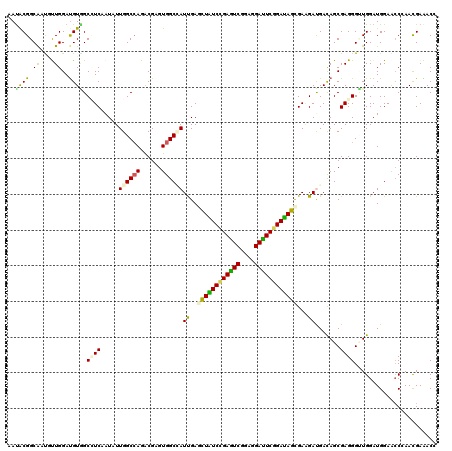
| Location | 10,981,536 – 10,981,656 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10981536 120 - 22407834 UGUUUCGUUCGGUUCCAUCCAGCCCUCGCUGUCGUCUUCGCUAUCCGAAUCCUCGGACUCGGAUAAUUCGAUAGCCACACGUCUGGCCAAGAUUGAAGCCACAUCCAAUGGCGCCGCGUU .........((((((.(((..(((..((.(((.((..(((.(((((((.((....)).)))))))...)))..)).)))))...)))...))).)).((((.......)))))))).... ( -34.60) >DroVir_CAF1 7394 120 - 1 GGUCUCAUUGGGUUCCAUCCAUCCCUCGCUGUCAUCCUCGCUAUCCGAAUCUUCUGACUCCGAUAGCUCAAUGGCCACUCGCCUGGCCAAAAUCGAGGCUACAUCCAGCAUUGCCGUAUU (((......(((..........)))..((((....((((((((((.((.((....)).)).))))))....((((((......)))))).....)))).......))))...)))..... ( -36.70) >DroGri_CAF1 8455 120 - 1 GGUUUCGUUCGGUUCCAUCCAACCCUCGCUGUCAUCUUCACUAUCGGAGUCCUCCGACUCCGACAGCUCGAUGGCCACUCGCCUGGCCAAUAUUGAGGCCACAUCCAGCAUUGCCGUAUU (((.......((((......))))...((((.........((.((((((((....)))))))).))(((((((((((......))))))...)))))........))))...)))..... ( -38.10) >DroWil_CAF1 9367 120 - 1 GGUUUCAUUGGGUUCCAUCCAACCCUCGCUAUCAUCUUCACUAUCCGAAUCCUCUGAUUCGGAUAACUCAAUUGCCACCCGUCUUGCCAAUAUAGAUGCCACAUCCAAAAGUGCCGUAUU (((..(...(((((......)))))..((.(((........((((((((((....)))))))))).....((((.((.......)).))))...)))))...........).)))..... ( -25.50) >DroMoj_CAF1 7688 120 - 1 GGUUUCAUUGGUUUCCAUCCAACCCUCGCUGUCAUCUUCACUGUCGGAAUCUUCCGACUCCGACAGCUCAAUGGCCACUCGCCUGGCCAAAAUCGAUGCCACAUCCAACAUUGCCGUAUU (((....((((....((((........((((((.........((((((....))))))...))))))....((((((......)))))).....))))......))))....)))..... ( -35.80) >DroAna_CAF1 9365 120 - 1 UGUCUCGUUUGGCUCCAUCCACCCCUCGCUGUCGUCAUCGCUGUCCGAGUCCUCCGACUCGGACAGUUCAAUUGCCACGCGUCUGGCCAAGAUGGAGGCGACGUCCAAAGGUGCCGUAUU ..........(((.((.........(((((..((((...((((((((((((....))))))))))))......((((......))))...))))..)))))........)).)))..... ( -46.13) >consensus GGUUUCAUUGGGUUCCAUCCAACCCUCGCUGUCAUCUUCACUAUCCGAAUCCUCCGACUCCGACAGCUCAAUGGCCACUCGCCUGGCCAAAAUCGAGGCCACAUCCAACAGUGCCGUAUU ((((((..................................((((((((.((....)).)))))))).......((((......)))).......)))))).................... (-20.75 = -20.92 + 0.17)
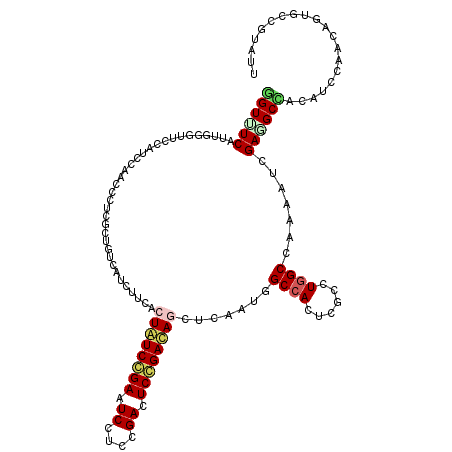
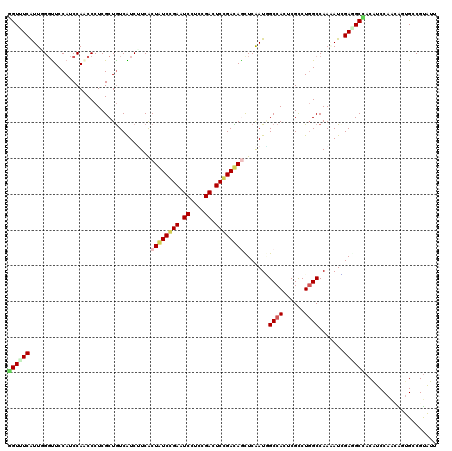
| Location | 10,981,576 – 10,981,696 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -46.25 |
| Consensus MFE | -29.40 |
| Energy contribution | -28.90 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
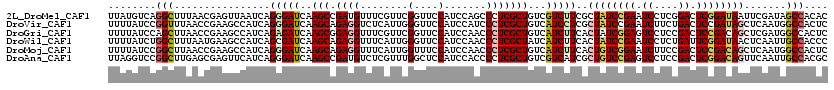
>2L_DroMel_CAF1 10981576 120 + 22407834 GUGUGGCUAUCGAAUUAUCCGAGUCCGAGGAUUCGGAUAGCGAAGACGACAGCGAGGGCUGGAUGGAACCGAACGAAACAUCGGCUUGAUCCCUGAUUAACUCGUUAAAGCCUGACAUAA ((((((((.(((...((((((((((....)))))))))).))).......((((((..(.((((.((.((((........)))).)).))))..).....))))))..))))..)))).. ( -42.90) >DroVir_CAF1 7434 120 + 1 GAGUGGCCAUUGAGCUAUCGGAGUCAGAAGAUUCGGAUAGCGAGGAUGACAGCGAGGGAUGGAUGGAACCCAAUGAGACCUCUGCUUGAUCCCUGAUGGCUUCGGUUAAACCGGAUAAAA .....((((((..((((((.(((((....))))).))))))..((((.(.(((((((..(.(.(((...))).).)..)))).)))).))))..))))))(((((.....)))))..... ( -46.20) >DroGri_CAF1 8495 120 + 1 GAGUGGCCAUCGAGCUGUCGGAGUCGGAGGACUCCGAUAGUGAAGAUGACAGCGAGGGUUGGAUGGAACCGAACGAAACCUCCGCUUGAUCUCUGAUGGCUUCGGUUAAGCUGGAUAAAA .....(((((((.((((((((((((....))))))))))))..((((.(.((((..(((((..((....))..)..))))..))))).)))).)))))))(((((.....)))))..... ( -48.70) >DroWil_CAF1 9407 120 + 1 GGGUGGCAAUUGAGUUAUCCGAAUCAGAGGAUUCGGAUAGUGAAGAUGAUAGCGAGGGUUGGAUGGAACCCAAUGAAACCUCUGCUUGAUCGCUGAUGGCUUCAUUAAAGCCAGAUAAAA ...((((..........((((((((....))))))))((((((((....(((((((((((......))))).........((.....))))))))....))))))))..))))....... ( -40.40) >DroMoj_CAF1 7728 120 + 1 GAGUGGCCAUUGAGCUGUCGGAGUCGGAAGAUUCCGACAGUGAAGAUGACAGCGAGGGUUGGAUGGAAACCAAUGAAACCUCUGCUUGAUCCCUGAUGGCUUCGGUUAAGCCGGAUAAAA .....((((((..((((((((((((....))))))))))))...(((.(.((((((((((((.(....))))))....)))).)))).)))...))))))(((((.....)))))..... ( -50.60) >DroAna_CAF1 9405 120 + 1 GCGUGGCAAUUGAACUGUCCGAGUCGGAGGACUCGGACAGCGAUGACGACAGCGAGGGGUGGAUGGAGCCAAACGAGACAUCGGCUUGAUCCCUGAUGAACUCGCUCAAGCCGGACCUAA ...((((.((((..(((((((((((....)))))))))))))))......((((((.((.((((.(((((............))))).))))))......))))))...))))....... ( -48.70) >consensus GAGUGGCCAUUGAGCUAUCCGAGUCGGAGGAUUCGGAUAGCGAAGAUGACAGCGAGGGUUGGAUGGAACCCAACGAAACCUCUGCUUGAUCCCUGAUGGCUUCGGUUAAGCCGGAUAAAA ....(((((((..((((((((((((....))))))))))))...))))..(((((((..((...(....)...))...)))).))).......................)))........ (-29.40 = -28.90 + -0.50)

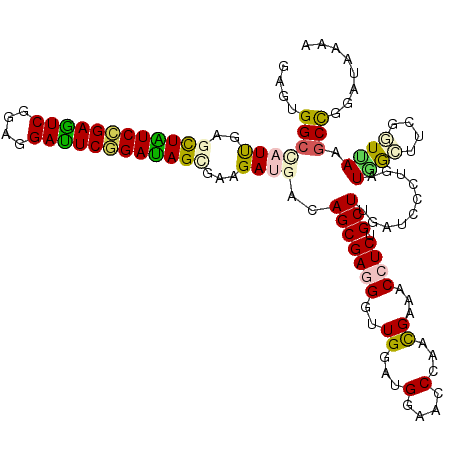
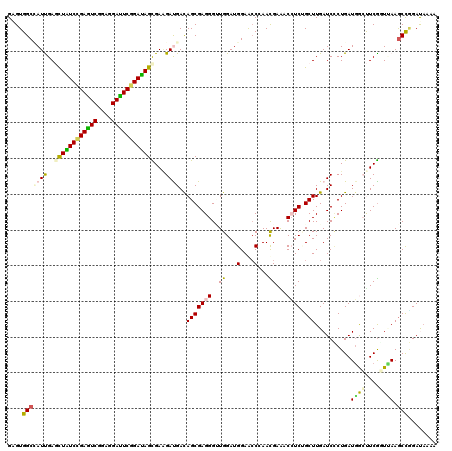
| Location | 10,981,576 – 10,981,696 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10981576 120 - 22407834 UUAUGUCAGGCUUUAACGAGUUAAUCAGGGAUCAAGCCGAUGUUUCGUUCGGUUCCAUCCAGCCCUCGCUGUCGUCUUCGCUAUCCGAAUCCUCGGACUCGGAUAAUUCGAUAGCCACAC ........((((.....((.....))..((((..((((((........))))))..))))))))...(((((((.......(((((((.((....)).)))))))...)))))))..... ( -38.10) >DroVir_CAF1 7434 120 - 1 UUUUAUCCGGUUUAACCGAAGCCAUCAGGGAUCAAGCAGAGGUCUCAUUGGGUUCCAUCCAUCCCUCGCUGUCAUCCUCGCUAUCCGAAUCUUCUGACUCCGAUAGCUCAAUGGCCACUC .......(((.....)))..(((((..(((((..(((.((((......(((((...)))))..)))))))...))))).((((((.((.((....)).)).))))))...)))))..... ( -36.40) >DroGri_CAF1 8495 120 - 1 UUUUAUCCAGCUUAACCGAAGCCAUCAGAGAUCAAGCGGAGGUUUCGUUCGGUUCCAUCCAACCCUCGCUGUCAUCUUCACUAUCGGAGUCCUCCGACUCCGACAGCUCGAUGGCCACUC ....................((((((.(((((..((((..((((......((......))))))..))))...)))))..((.((((((((....)))))))).))...))))))..... ( -39.00) >DroWil_CAF1 9407 120 - 1 UUUUAUCUGGCUUUAAUGAAGCCAUCAGCGAUCAAGCAGAGGUUUCAUUGGGUUCCAUCCAACCCUCGCUAUCAUCUUCACUAUCCGAAUCCUCUGAUUCGGAUAACUCAAUUGCCACCC .......((((...(((((((((.((.((......)).)))))))))))(((((......)))))................((((((((((....))))))))))........))))... ( -37.60) >DroMoj_CAF1 7728 120 - 1 UUUUAUCCGGCUUAACCGAAGCCAUCAGGGAUCAAGCAGAGGUUUCAUUGGUUUCCAUCCAACCCUCGCUGUCAUCUUCACUGUCGGAAUCUUCCGACUCCGACAGCUCAAUGGCCACUC .......(((.....)))..(((((..(((((..(((.((((.....((((.......)))).)))))))...)))....((((((((.((....)).))))))))))..)))))..... ( -36.10) >DroAna_CAF1 9405 120 - 1 UUAGGUCCGGCUUGAGCGAGUUCAUCAGGGAUCAAGCCGAUGUCUCGUUUGGCUCCAUCCACCCCUCGCUGUCGUCAUCGCUGUCCGAGUCCUCCGACUCGGACAGUUCAAUUGCCACGC ...(((.((((...((((((........((((..((((((........))))))..))))....)))))))))).....((((((((((((....))))))))))))......))).... ( -48.10) >consensus UUUUAUCCGGCUUAACCGAAGCCAUCAGGGAUCAAGCAGAGGUUUCAUUGGGUUCCAUCCAACCCUCGCUGUCAUCUUCACUAUCCGAAUCCUCCGACUCCGACAGCUCAAUGGCCACUC ........(((................(((((..(((.((((........(....).......)))))))...)))))..((((((((.((....)).)))))))).......))).... (-21.42 = -21.73 + 0.31)
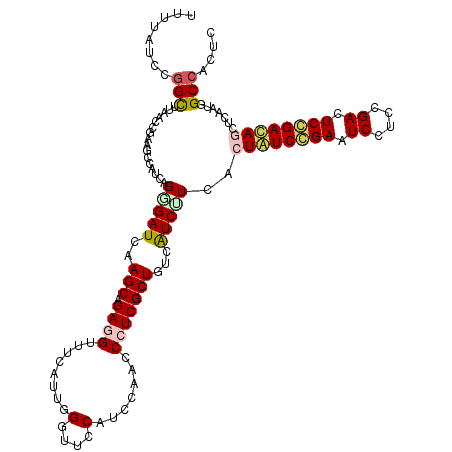
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:42 2006