| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,979,894 – 10,980,090 |
| Length | 196 |
| Max. P | 0.974323 |

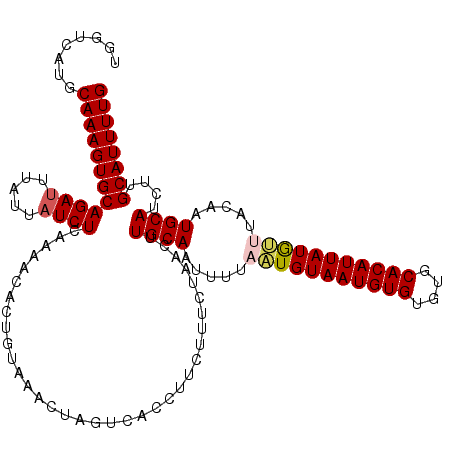
| Location | 10,979,894 – 10,980,010 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.64 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10979894 116 + 22407834 UGGUCAUGCAAAGUGCAGAUUUAUUAUCUCAAAACACUGUAAACUAGUCACCUU----CUAACUUGCAAUUUUAAUGUAAUGUGUGUGCACAUUAUAUUUACAAUGCAUCUUGCAUUUUG .....((((((..((((....................(((((..(((.......----)))..))))).....(((((((((((....))))))))))).....))))..)))))).... ( -26.80) >DroSec_CAF1 8087 120 + 1 UGGUCAUGCAAAGUGCAGAUUAAUUAUCUCAAAACACUGUAAACUAGUCACCUUCUUUCUAACUUGCAAUUUGAAUGUAAUGUGUGUGCACAUUAUGUUUACGAUGCAUCUGGCAUUUUG ........((((((((((((....((((.........(((((..(((...........)))..)))))...(((((((((((((....))))))))))))).)))).)))).)))))))) ( -27.10) >DroSim_CAF1 7359 120 + 1 UGGUCAUGCAAAGUGCAGAAUUAUUAUCUCAAAACACUGUAAACUAGUCACCUUCUUUCUAACUUGCACUUUGAAUGUAAUGUGUGUGCACAUUAUGUUUACGAUGCAUCUGGCAUUUUG ........(((((((((((................((((.....))))................((((.(.(((((((((((((....))))))))))))).).))))))).)))))))) ( -26.80) >DroEre_CAF1 8077 119 + 1 -GGUCAUGCAAAGUGCAGAUUUAUUAUCUCAAAACACUGUAAACUAGUCAACUUCUUUCUACCUUGCAAUAUUUUUGUAAUGUGUGUGCACAUUAUGAUUACAAUGCAUUUUGCAUUUUG -....((((((((((((....................(((((..(((...........)))..)))))......(..(((((((....)))))))..)......)))))))))))).... ( -28.00) >DroYak_CAF1 7405 115 + 1 -GGUCAUGCAAAGUGCAGAUUUUUUAUCUCAAAACACUGUUAACUAUUCACCU----UCUACCUUGCAAUAUUUAUGUAAUGUGUGUGCACAUUAUGGUUAUAAUGCAGUUUGCAUUUUG -.......((((((((((((.........((......))..............----.......((((.(((...(((((((((....)))))))))...))).)))))))))))))))) ( -24.80) >consensus UGGUCAUGCAAAGUGCAGAUUUAUUAUCUCAAAACACUGUAAACUAGUCACCUUCUUUCUAACUUGCAAUUUUAAUGUAAUGUGUGUGCACAUUAUGUUUACAAUGCAUCUUGCAUUUUG ........((((((((((((.....))))...................................((((.....(((((((((((....))))))))))).....))))....)))))))) (-22.24 = -22.64 + 0.40)


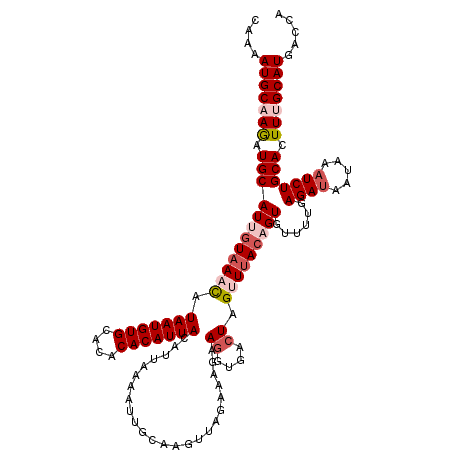
| Location | 10,979,894 – 10,980,010 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -20.68 |
| Energy contribution | -22.28 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10979894 116 - 22407834 CAAAAUGCAAGAUGCAUUGUAAAUAUAAUGUGCACACACAUUACAUUAAAAUUGCAAGUUAG----AAGGUGACUAGUUUACAGUGUUUUGAGAUAAUAAAUCUGCACUUUGCAUGACCA ....(((((((.((((((((((((.(((((((....)))))))...................----.((....)).)))))))))......((((.....))))))).)))))))..... ( -27.80) >DroSec_CAF1 8087 120 - 1 CAAAAUGCCAGAUGCAUCGUAAACAUAAUGUGCACACACAUUACAUUCAAAUUGCAAGUUAGAAAGAAGGUGACUAGUUUACAGUGUUUUGAGAUAAUUAAUCUGCACUUUGCAUGACCA ((((.(((((((.((((.((((((.(((((((....)))))))........................((....)).)))))).))))))))((((.....))))))).))))........ ( -25.20) >DroSim_CAF1 7359 120 - 1 CAAAAUGCCAGAUGCAUCGUAAACAUAAUGUGCACACACAUUACAUUCAAAGUGCAAGUUAGAAAGAAGGUGACUAGUUUACAGUGUUUUGAGAUAAUAAUUCUGCACUUUGCAUGACCA ....((((.....))))........(((((((....)))))))(((.((((((((((((((......((....)).((((.(((....)))))))..))))).))))))))).))).... ( -27.00) >DroEre_CAF1 8077 119 - 1 CAAAAUGCAAAAUGCAUUGUAAUCAUAAUGUGCACACACAUUACAAAAAUAUUGCAAGGUAGAAAGAAGUUGACUAGUUUACAGUGUUUUGAGAUAAUAAAUCUGCACUUUGCAUGACC- ....(((((((.((((((((.(((.(((((((....)))))))..(((((((((.(((.(((...........))).))).)))))))))..))).))))...)))).)))))))....- ( -29.00) >DroYak_CAF1 7405 115 - 1 CAAAAUGCAAACUGCAUUAUAACCAUAAUGUGCACACACAUUACAUAAAUAUUGCAAGGUAGA----AGGUGAAUAGUUAACAGUGUUUUGAGAUAAAAAAUCUGCACUUUGCAUGACC- ....(((((((.(((..........(((((((....)))))))((.((((((((..((.((..----.......)).))..))))))))))((((.....))))))).)))))))....- ( -22.30) >consensus CAAAAUGCAAGAUGCAUUGUAAACAUAAUGUGCACACACAUUACAUUAAAAUUGCAAGUUAGAAAGAAGGUGACUAGUUUACAGUGUUUUGAGAUAAUAAAUCUGCACUUUGCAUGACCA ....(((((((.((((((((((((.(((((((....)))))))........................((....)).)))))))))......((((.....))))))).)))))))..... (-20.68 = -22.28 + 1.60)


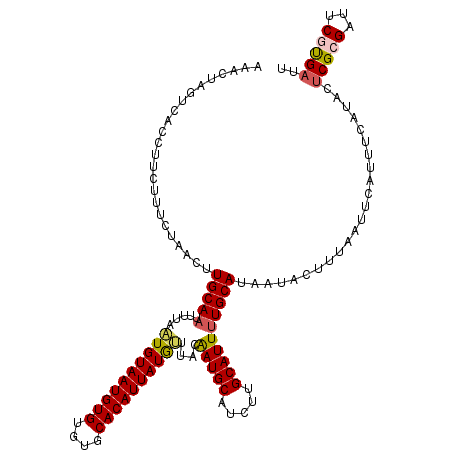
| Location | 10,979,934 – 10,980,050 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.06 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10979934 116 + 22407834 AAACUAGUCACCUU----CUAACUUGCAAUUUUAAUGUAAUGUGUGUGCACAUUAUAUUUACAAUGCAUCUUGCAUUUUGCAUAAUACUUUAAUUCAUUUCAUACUCGCGAUUCGUGAUU .....((((((..(----(.....((((.....(((((((((((....))))))))))).....))))...(((.....)))...........................))...)))))) ( -22.80) >DroSec_CAF1 8127 120 + 1 AAACUAGUCACCUUCUUUCUAACUUGCAAUUUGAAUGUAAUGUGUGUGCACAUUAUGUUUACGAUGCAUCUGGCAUUUUGCAUAAUACUUUAAUUCAUUUCAUACUCGCGAUUCGUGAUU .....((((((..((.........(((((..(((((((((((((....))))))))))))).(((((.....))))))))))(((....))).................))...)))))) ( -24.00) >DroSim_CAF1 7399 120 + 1 AAACUAGUCACCUUCUUUCUAACUUGCACUUUGAAUGUAAUGUGUGUGCACAUUAUGUUUACGAUGCAUCUGGCAUUUUGCAUAAUACUUUAAUUCAUUUCAUACUCGCGAUUCGUGAUU .....((((((..((.........((((.(.(((((((((((((....))))))))))))).).))))....((.....))............................))...)))))) ( -23.20) >DroEre_CAF1 8116 120 + 1 AAACUAGUCAACUUCUUUCUACCUUGCAAUAUUUUUGUAAUGUGUGUGCACAUUAUGAUUACAAUGCAUUUUGCAUUUUGCAUUAUACUUAAAUUCAUUUCAUACACGCGAUUCAUGAUU .....(((((.............((((((.....))))))((((((((......((((.......(.((..(((.....)))..)).)......))))....)))))))).....))))) ( -18.42) >DroYak_CAF1 7444 116 + 1 UAACUAUUCACCU----UCUACCUUGCAAUAUUUAUGUAAUGUGUGUGCACAUUAUGGUUAUAAUGCAGUUUGCAUUUUGCAUAAUACUUAAAUUCAUUUCAUACUCGCGAUUCACGAUU .............----......((((..(((...(((((((((....)))))))))...)))((((((........))))))........................))))......... ( -17.80) >consensus AAACUAGUCACCUUCUUUCUAACUUGCAAUUUUAAUGUAAUGUGUGUGCACAUUAUGUUUACAAUGCAUCUUGCAUUUUGCAUAAUACUUUAAUUCAUUUCAUACUCGCGAUUCGUGAUU ........................(((((.....((((((((((....))))))))))....(((((.....)))))))))).......................(((((...))))).. (-16.96 = -17.16 + 0.20)

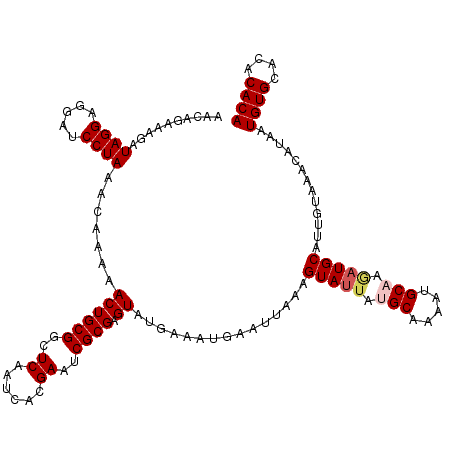
| Location | 10,979,970 – 10,980,090 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -21.41 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10979970 120 - 22407834 AACAGAAAGAUAGGAGGAUCCUAAACAAAAACUGCGGCUCAAUCACGAAUCGCGAGUAUGAAAUGAAUUAAAGUAUUAUGCAAAAUGCAAGAUGCAUUGUAAAUAUAAUGUGCACACACA ..........((((.....)))).((((..((((((..((......))..))).)))...............(((((.(((.....))).))))).))))........((((....)))) ( -23.20) >DroSec_CAF1 8167 120 - 1 AACAGAAAGAUAGGAGGAUCCUAAAGAAAAACUGCGGCUCAAUCACGAAUCGCGAGUAUGAAAUGAAUUAAAGUAUUAUGCAAAAUGCCAGAUGCAUCGUAAACAUAAUGUGCACACACA .((.......((((.....))))..((...((((((..((......))..))).)))...............(((((..((.....))..))))).))))........((((....)))) ( -18.80) >DroSim_CAF1 7439 120 - 1 AACAGAAAGAUAGGAGGAUCCUAAACAAAAACUGCGGCUCAAUCACGAAUCGCGAGUAUGAAAUGAAUUAAAGUAUUAUGCAAAAUGCCAGAUGCAUCGUAAACAUAAUGUGCACACACA ..........((((.....)))).((....((((((..((......))..))).)))...............(((((..((.....))..)))))...))........((((....)))) ( -18.50) >DroEre_CAF1 8156 120 - 1 AACAGAAAGAUAGGAGGAUCCUAAACAAAAACUGCGGCUCAAUCAUGAAUCGCGUGUAUGAAAUGAAUUUAAGUAUAAUGCAAAAUGCAAAAUGCAUUGUAAUCAUAAUGUGCACACACA ..........((((.....))))..........(((..(((....)))..)))(((((((..((((.......(((((((((..........))))))))).))))..)))))))..... ( -26.50) >DroYak_CAF1 7480 120 - 1 AACAGAAAGAUAGGAGGAUCCUAAACAAAAACUGCGGAUCAAUCGUGAAUCGCGAGUAUGAAAUGAAUUUAAGUAUUAUGCAAAAUGCAAACUGCAUUAUAACCAUAAUGUGCACACACA ............((..(((((..............)))))..(((((...)))))((((((.((........)).))))))..(((((.....)))))....))....((((....)))) ( -20.04) >consensus AACAGAAAGAUAGGAGGAUCCUAAACAAAAACUGCGGCUCAAUCACGAAUCGCGAGUAUGAAAUGAAUUAAAGUAUUAUGCAAAAUGCAAGAUGCAUUGUAAACAUAAUGUGCACACACA ..........((((.....)))).......((((((..((......))..)))).))...............(((((.(((.....))).))))).............((((....)))) (-18.42 = -19.10 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:37 2006