
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,963,792 – 10,963,971 |
| Length | 179 |
| Max. P | 0.988543 |

| Location | 10,963,792 – 10,963,902 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 66.91 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10963792 110 + 22407834 UCCCAUGAAUUUAACCAUUUAAACAUCCAA----UGUUAGUUUUUAACAGACAAUUAAUUCAG-GGGACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUC ((((.((((((..........(((((...)----)))).((((.....))))....)))))).-))))..............((((((.....))))))................ ( -24.50) >DroSec_CAF1 46554 97 + 1 UCCCAUAAAUUAAACCAUUUAAACAUCCAA----AGUUAGUUUUUAACAGACAAUUGAUU--------------CUUUCACUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUC .............................(----((((.((((.....))))..((((.(--------------(.......((((((.....))))))...)).))))))))). ( -17.40) >DroSim_CAF1 53467 97 + 1 UCCCAUAAAUUAAACCAUUUAAACAUCCAA----UGUUAGUUUUUAACAGACAAUUGAUU--------------CUUUCACUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUC ...........................(((----((((.((.....)).))).)))).((--------------(..((...((((((.....))))))...))...)))..... ( -17.70) >DroEre_CAF1 47135 97 + 1 UU-CAUAAACUGAAG-------------AA----UGUCAGUAUUUUGUUGGUGGUUGAUGCAAGGGGACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUC ((-(....(((((..-------------..----..))))).....((((((((..((((...(....)..))))..)))).((((((.....))))))..))))..)))..... ( -27.50) >DroYak_CAF1 48771 110 + 1 UC-CAUAAACGGAAGUAUUCAAGCAUCCAA----UGUUAGUAUUUGGUUGGUUGUUGAUACAGGGGGGCAAUAUCUUUCACUGGCCGCUUAAAGCGGCCUACAAUUCGAAACUUC ..-......((((.((((((((.((.((((----.........)))).)).)))..))))).(..(((.....)))..)...((((((.....)))))).....))))....... ( -29.60) >DroAna_CAF1 61430 95 + 1 UC-CAUUAAUUUAAUU-UUAAGACUUCCCACUGCAAUAAGUAUUUC-----------UGUCUA-GCGACAAUAU------CUGGCCGCGUAAAGCGGCCACAAUUUGAAAACUGC ..-.............-...((((.....(((......))).....-----------.)))).-....(((...------.(((((((.....)))))))....)))........ ( -20.50) >consensus UC_CAUAAAUUAAACCAUUUAAACAUCCAA____UGUUAGUAUUUAACAGACAAUUGAUUCA__GGGACAAUAUCUUUCACUGGCCGCUUAAAGCGGCCUCCAAUUCGAAACUUC ..................................................................................((((((.....))))))................ (-13.20 = -13.20 + 0.00)


| Location | 10,963,792 – 10,963,902 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.91 |
| Mean single sequence MFE | -23.89 |
| Consensus MFE | -13.82 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10963792 110 - 22407834 GAAGUUUCGAAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUCCC-CUGAAUUAAUUGUCUGUUAAAAACUAACA----UUGGAUGUUUAAAUGGUUAAAUUCAUGGGA ................((((((.....))))))..............((((-.(((((((((..(...(((((..(((...----.)))...))))))..))).)))))).)))) ( -27.10) >DroSec_CAF1 46554 97 - 1 GAAGUUUCGAAUCGGAGGCCGCUUUAAGCGGCCAGUGAAAG--------------AAUCAAUUGUCUGUUAAAAACUAACU----UUGGAUGUUUAAAUGGUUUAAUUUAUGGGA ..(((((..((.((((((((((.....))))))..(((...--------------..)))....))))))..)))))..((----.(((((..............))))).)).. ( -20.54) >DroSim_CAF1 53467 97 - 1 GAAGUUUCGAAUCGGAGGCCGCUUUAAGCGGCCAGUGAAAG--------------AAUCAAUUGUCUGUUAAAAACUAACA----UUGGAUGUUUAAAUGGUUUAAUUUAUGGGA .(((((..((((((..((((((.....))))))..((((..--------------.(((...(((..((.....))..)))----...))).))))..)))))))))))...... ( -21.80) >DroEre_CAF1 47135 97 - 1 GAAGUUUCGAAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUCCCCUUGCAUCAACCACCAACAAAAUACUGACA----UU-------------CUUCAGUUUAUG-AA ...........((((.((((((.....)))))).(..(..(((...)))...)..)........))))((....(((((..----..-------------..)))))...))-.. ( -22.00) >DroYak_CAF1 48771 110 - 1 GAAGUUUCGAAUUGUAGGCCGCUUUAAGCGGCCAGUGAAAGAUAUUGCCCCCCUGUAUCAACAACCAACCAAAUACUAACA----UUGGAUGCUUGAAUACUUCCGUUUAUG-GA ((((((((((.((((.((((((.....)))))).......(((((.........))))).)))).((.((((.........----)))).)).))))).)))))........-.. ( -26.90) >DroAna_CAF1 61430 95 - 1 GCAGUUUUCAAAUUGUGGCCGCUUUACGCGGCCAG------AUAUUGUCGC-UAGACA-----------GAAAUACUUAUUGCAGUGGGAAGUCUUAA-AAUUAAAUUAAUG-GA ..((.((((..(((((((((((.....))))))..------...(((((..-..))))-----------)...........)))))..)))).))...-.............-.. ( -25.00) >consensus GAAGUUUCGAAUUGGAGGCCGCUUUAAGCGGCCAGUGAAAGAUAUUGUCCC__UGAAUCAAUUGUCUGUUAAAAACUAACA____UUGGAUGUUUAAAUGGUUAAAUUUAUG_GA ................((((((.....)))))).................................................................................. (-13.82 = -13.82 + 0.00)

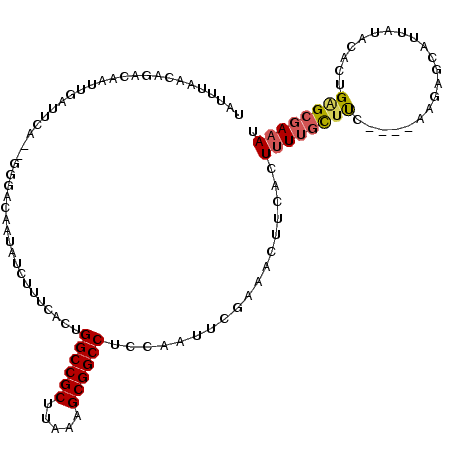
| Location | 10,963,828 – 10,963,938 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.31 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -15.09 |
| Energy contribution | -15.37 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10963828 110 + 22407834 UUUUUAACAGACAAUUAAUUCAG-GGGACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCCUC----AAGAGCAUUAUACACUGAGCGAAAU ..................(((.(-(((.(((...........((((((.....))))))........((....))....)))))))----....((.(((.....)))))))).. ( -21.00) >DroSec_CAF1 46590 97 + 1 UUUUUAACAGACAAUUGAUU--------------CUUUCACUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUCACUUUUGCUUC----AAGAGCAUUAUACACUGAGCGAAAU .......(((....((((.(--------------(.......((((((.....))))))...)).))))...........(((((.----..))))).......)))........ ( -20.80) >DroSim_CAF1 53503 97 + 1 UUUUUAACAGACAAUUGAUU--------------CUUUCACUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUCACUUUUGCUCC----AAGAGCAUUAUACACUGAGCGAAAU .......(((....((((.(--------------(.......((((((.....))))))...)).))))...........(((((.----..))))).......)))........ ( -22.40) >DroEre_CAF1 47157 111 + 1 UAUUUUGUUGGUGGUUGAUGCAAGGGGACAAUAUCUUUCACUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCUUC----GCGAAUAUUAUACACUGAGCGAAAU .((((((((((((.(((..(((((((((..............((((((.....))))))..............)).)))))))...----.)))........)))).)))))))) ( -29.19) >DroYak_CAF1 48806 111 + 1 UAUUUGGUUGGUUGUUGAUACAGGGGGGCAAUAUCUUUCACUGGCCGCUUAAAGCGGCCUACAAUUCGAAACUUCACUGUAGCUUC----AAGAGCCUUAUACACUGAGCGAAAU ..((..((.((((.((((((((((..((......))..)...((((((.....)))))).................)))))...))----)).))))......))..))...... ( -30.30) >DroAna_CAF1 61468 97 + 1 UAUUUC-----------UGUCUA-GCGACAAUAU------CUGGCCGCGUAAAGCGGCCACAAUUUGAAAACUGCACUUUUCUUUUUUUUUUUACCACAUUACACUGGGAGAAAG ..((((-----------(.((((-(.(.......------.(((((((.....)))))))......(((((......)))))....................).)))))))))). ( -24.20) >consensus UAUUUAACAGACAAUUGAUUCA__GGGACAAUAUCUUUCACUGGCCGCUUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCUUC____AAGAGCAUUAUACACUGAGCGAAAU ..........................................((((((.....))))))..................((((((((.....................)))))))). (-15.09 = -15.37 + 0.28)

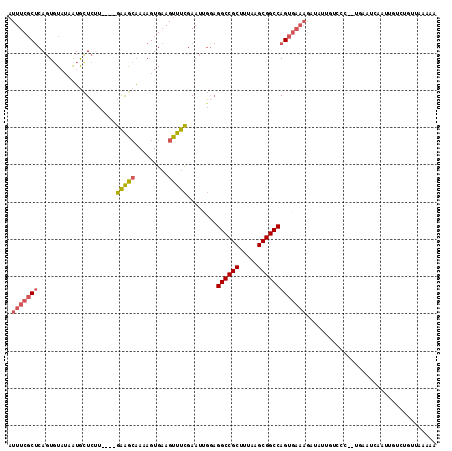
| Location | 10,963,828 – 10,963,938 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.31 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10963828 110 - 22407834 AUUUCGCUCAGUGUAUAAUGCUCUU----GAGGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUCCC-CUGAAUUAAUUGUCUGUUAAAAA .......((((.(.((((((.((((----(((((.........)))))........((((((.....)))))).....)))))))))).).-)))).(((((.....)))))... ( -26.90) >DroSec_CAF1 46590 97 - 1 AUUUCGCUCAGUGUAUAAUGCUCUU----GAAGCAAAAGUGAAGUUUCGAAUCGGAGGCCGCUUUAAGCGGCCAGUGAAAG--------------AAUCAAUUGUCUGUUAAAAA .(((((((...............((----(((((.........)))))))......((((((.....))))))))))))).--------------.................... ( -24.50) >DroSim_CAF1 53503 97 - 1 AUUUCGCUCAGUGUAUAAUGCUCUU----GGAGCAAAAGUGAAGUUUCGAAUCGGAGGCCGCUUUAAGCGGCCAGUGAAAG--------------AAUCAAUUGUCUGUUAAAAA .(((((((..........(((((..----.))))).....................((((((.....))))))))))))).--------------.................... ( -24.80) >DroEre_CAF1 47157 111 - 1 AUUUCGCUCAGUGUAUAAUAUUCGC----GAAGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAUGCGGCCAGUGAAAGAUAUUGUCCCCUUGCAUCAACCACCAACAAAAUA .....((..((.(.(((((((((((----(((((.........)))))........((((((.....)))))).))))...))))))).).)).))................... ( -27.20) >DroYak_CAF1 48806 111 - 1 AUUUCGCUCAGUGUAUAAGGCUCUU----GAAGCUACAGUGAAGUUUCGAAUUGUAGGCCGCUUUAAGCGGCCAGUGAAAGAUAUUGCCCCCCUGUAUCAACAACCAACCAAAUA .(((((((...........((..((----((((((.......))))))))...)).((((((.....)))))))))))))(((((.........)))))................ ( -29.00) >DroAna_CAF1 61468 97 - 1 CUUUCUCCCAGUGUAAUGUGGUAAAAAAAAAAAGAAAAGUGCAGUUUUCAAAUUGUGGCCGCUUUACGCGGCCAG------AUAUUGUCGC-UAGACA-----------GAAAUA .(((((..(((((((((((..............(((((......)))))......(((((((.....))))))).------)))))).)))-).)..)-----------)))).. ( -25.10) >consensus AUUUCGCUCAGUGUAUAAUGCUCUU____GAAGCAAAAGUGAAGUUUCGAAUUGGAGGCCGCUUUAAGCGGCCAGUGAAAGAUAUUGUCCC__UGAAUCAAUUGUCUGUUAAAAA .(((((((.....................(((((.........)))))........((((((.....)))))))))))))................................... (-19.41 = -19.88 + 0.48)


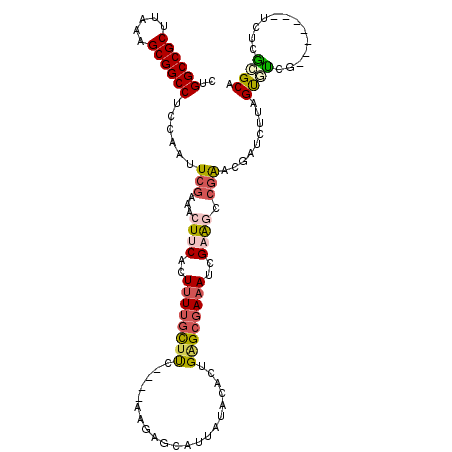
| Location | 10,963,867 – 10,963,971 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -17.73 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10963867 104 + 22407834 CUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCCUC----AAGAGCAUUAUACACUGAGCGAAAUCGAAGCCGAACGAUCUUAGUGUCG-------UCUCGCGCA ..((((((.....)))))).....(((((........((((((.((----(..............))))))))))))))((((((((((......))))-------).))).)). ( -29.94) >DroSec_CAF1 46616 104 + 1 CUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUCACUUUUGCUUC----AAGAGCAUUAUACACUGAGCGAAAUCGAAACCGAACGAUGUUAGUAUCG-------UCUCGCGCA ..((((((.....))))))..(((((((............(((((.----..)))))............)).)))))....(((((((((....)))))-------).))).... ( -29.15) >DroSim_CAF1 53529 104 + 1 CUGGCCGCUUAAAGCGGCCUCCGAUUCGAAACUUCACUUUUGCUCC----AAGAGCAUUAUACACUGAGCGAAAUCGAAGCCGAACGAUGUUAGUAUCG-------UCUCGCGCA ..((((((.....))))))..(((((((............(((((.----..)))))............)).)))))..(((((((((((....)))))-------).))).)). ( -34.85) >DroEre_CAF1 47197 102 + 1 CUGGCCGCAUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCUUC----GCGAAUAUUAUACACUGAGCGAAAUUGCAGCCGAUCGAUCUUAGUGUCG-------UC--ACGCA ..((((((.....))))))......((((..((.((.((((((((.----(.(.........).).)))))))).)).))....)))).....((((..-------..--)))). ( -25.90) >DroYak_CAF1 48846 102 + 1 CUGGCCGCUUAAAGCGGCCUACAAUUCGAAACUUCACUGUAGCUUC----AAGAGCCUUAUACACUGAGCGAAAUCGAAGUCGAUCGACCUUAGUGCCG-------UC--GCGCA ..((((((.....)))))).....(((((..(((((.((((((((.----..))))....)))).)))).)...)))))(((....)))....((((..-------..--)))). ( -29.10) >DroAna_CAF1 61490 112 + 1 CUGGCCGCGUAAAGCGGCCACAAUUUGAAAACUGCACUUUUCUUUUUUUUUUUACCACAUUACACUGGGAGAAAGGGAGACCGGU---UCCUUGCUUCUUCGUCAGUGUUAAGCA .(((((((.....)))))))......(((((......)))))...................((((((((((((((((((.....)---)))))..))))))..))))))...... ( -31.50) >consensus CUGGCCGCUUAAAGCGGCCUCCAAUUCGAAACUUCACUUUUGCUUC____AAGAGCAUUAUACACUGAGCGAAAUCGAAGCCGAACGAUCUUAGUGUCG_______UCUCGCGCA ..((((((.....))))))......(((...((((..((((((((.....................))))))))..)))).))).........((((.............)))). (-17.73 = -18.40 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:32 2006