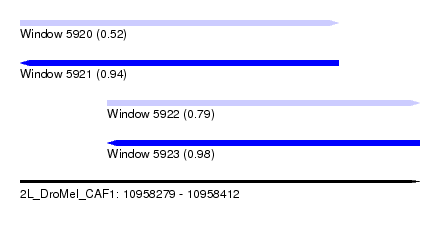
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,958,279 – 10,958,412 |
| Length | 133 |
| Max. P | 0.976029 |

| Location | 10,958,279 – 10,958,385 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.46 |
| Mean single sequence MFE | -24.49 |
| Consensus MFE | -12.35 |
| Energy contribution | -12.55 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
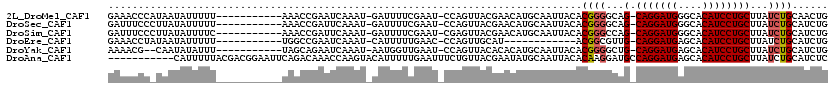
>2L_DroMel_CAF1 10958279 106 + 22407834 GAAACCCAUAAUAUUUUU-----------AAACCGAAUCAAAU-GAUUUUCGAAU-CCAGUUACGAACAUGCAAUUACACGGGGCAG-CAGGAUGGGCACAUCCUGCUUAUCUGCAACUG ....(((...........-----------.....(((((....-)))))((((((-...))).)))..............)))((((-(((((((....)))))))).....)))..... ( -23.60) >DroSec_CAF1 41238 106 + 1 GAUUUCCCUUAUAUUUUU-----------AAACCGAUUCAAAU-GAUUUUCGAAU-CCAGUUACGAACAUGCAAUUACACGGGGCAG-CAGGAUGGGCACAUCCUGCUUAUCUGCAUCUG .....(((..........-----------.....((((((((.-...))).))))-)..((.....))............)))((((-(((((((....)))))))).....)))..... ( -23.20) >DroSim_CAF1 48214 106 + 1 GAUUUCCCUUAUAUUUUC-----------AAACCGAUUCAAAU-GAUUUUCGAAU-CGAGUUACGAACAUGCAAUUACACGGGCCAG-CAGGAUGGGCACAUCCUGCUUAUCUGCAUCUG ...........(((.(((-----------.((((((((((((.-...))).))))-)).)))..))).)))........((((..((-(((((((....)))))))))..))))...... ( -27.20) >DroEre_CAF1 41960 94 + 1 GAAACCUAUAAUAUUUUU-----------UGGCCGAAUCAAAU-CAUUUUUGAAC-CCAGUUGCAU------------ACGGCGUUG-CAGGAUGAGCACAUCCUGCUUAUCUGCAUCUG ..................-----------..(((...(((((.-....)))))..-...((.....------------)))))(.((-(((.(((((((.....)))))))))))).).. ( -21.90) >DroYak_CAF1 43390 104 + 1 AAAACG--CAAUAUAUUU-----------UAGCAGAAUCAAAU-AAUGGUUGAAU-CCAGUUACACACAUGCAAUUACACGGGGCUG-CAGGAUGAGCACAUCCUGCUUAUCUGCAUCUG .....(--(.........-----------..(((.........-..(((......-)))..........)))........(((...(-(((((((....))))))))...)))))..... ( -23.60) >DroAna_CAF1 49359 109 + 1 -----------CAUUUUUACGACGGAAUUCAGACAAACCAAGUACAUUUUUGAAUUUCUGUUACGAAUAUGCAAUUACACAAGGAUGCCAGGAUGAGCACAUCCUGCUUAUCUGCAUCUC -----------(((.(((..(((((((((((((...............)))))).)))))))..))).)))...........((((((..(((((((((.....))))))))))))))). ( -27.46) >consensus GAAACCC_UAAUAUUUUU___________AAACCGAAUCAAAU_GAUUUUCGAAU_CCAGUUACGAACAUGCAAUUACACGGGGCAG_CAGGAUGAGCACAUCCUGCUUAUCUGCAUCUG ...............................................................................((((...(.(((((((....))))))))...))))...... (-12.35 = -12.55 + 0.20)

| Location | 10,958,279 – 10,958,385 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.46 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -15.56 |
| Energy contribution | -16.23 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10958279 106 - 22407834 CAGUUGCAGAUAAGCAGGAUGUGCCCAUCCUG-CUGCCCCGUGUAAUUGCAUGUUCGUAACUGG-AUUCGAAAAUC-AUUUGAUUCGGUUU-----------AAAAAUAUUAUGGGUUUC (((((((.....(((((((((....)))))))-))....(((((....)))))...)))))))(-(.(((((....-.))))).)).....-----------.................. ( -30.80) >DroSec_CAF1 41238 106 - 1 CAGAUGCAGAUAAGCAGGAUGUGCCCAUCCUG-CUGCCCCGUGUAAUUGCAUGUUCGUAACUGG-AUUCGAAAAUC-AUUUGAAUCGGUUU-----------AAAAAUAUAAGGGAAAUC ............(((((((((....)))))))-))..(((((......))(((((..(((.(.(-(((((((....-.)))))))).).))-----------)..)))))..)))..... ( -30.00) >DroSim_CAF1 48214 106 - 1 CAGAUGCAGAUAAGCAGGAUGUGCCCAUCCUG-CUGGCCCGUGUAAUUGCAUGUUCGUAACUCG-AUUCGAAAAUC-AUUUGAAUCGGUUU-----------GAAAAUAUAAGGGAAAUC ............(((((((((....)))))))-))..(((((((....)))).((((.(((.((-(((((((....-.)))))))))))))-----------))).......)))..... ( -33.90) >DroEre_CAF1 41960 94 - 1 CAGAUGCAGAUAAGCAGGAUGUGCUCAUCCUG-CAACGCCGU------------AUGCAACUGG-GUUCAAAAAUG-AUUUGAUUCGGCCA-----------AAAAAUAUUAUAGGUUUC (((.((((.....((((((((....)))))))-)........------------.)))).)))(-(((((((....-.)))))....))).-----------.................. ( -23.84) >DroYak_CAF1 43390 104 - 1 CAGAUGCAGAUAAGCAGGAUGUGCUCAUCCUG-CAGCCCCGUGUAAUUGCAUGUGUGUAACUGG-AUUCAACCAUU-AUUUGAUUCUGCUA-----------AAAUAUAUUG--CGUUUU .((((((((....((((((((....)))))))-)(((..(((((....))))).........((-(.((((.....-..)))).)))))).-----------.......)))--))))). ( -29.80) >DroAna_CAF1 49359 109 - 1 GAGAUGCAGAUAAGCAGGAUGUGCUCAUCCUGGCAUCCUUGUGUAAUUGCAUAUUCGUAACAGAAAUUCAAAAAUGUACUUGGUUUGUCUGAAUUCCGUCGUAAAAAUG----------- ..(((((((((((((((((((....)))))).((((...(((((....)))))(((......)))........)))).....))))))))).....)))).........----------- ( -25.10) >consensus CAGAUGCAGAUAAGCAGGAUGUGCCCAUCCUG_CAGCCCCGUGUAAUUGCAUGUUCGUAACUGG_AUUCAAAAAUC_AUUUGAUUCGGCUU___________AAAAAUAUUA_GGGUUUC (((.(((......((((((((....))))))).).....(((((....)))))...))).)))....(((((......)))))..................................... (-15.56 = -16.23 + 0.67)
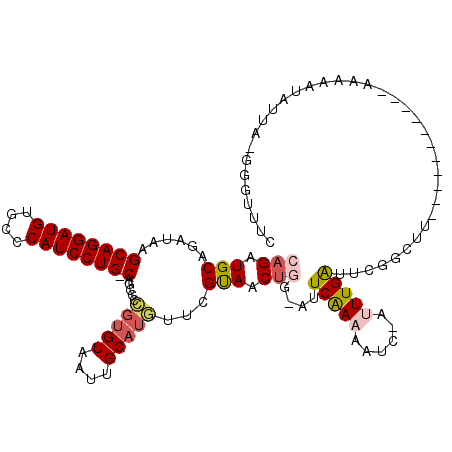
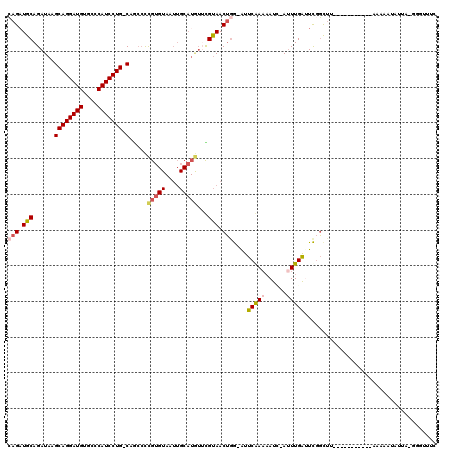
| Location | 10,958,308 – 10,958,412 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.55 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
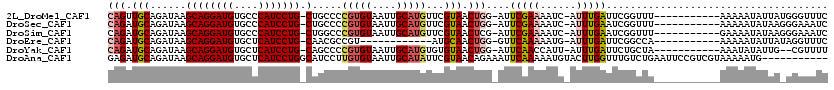
>2L_DroMel_CAF1 10958308 104 + 22407834 AAU-GAUUUUCGAAU-CCAGUUACGAACAUGCAAUUACACGGGGCAG-CAGGAUGGGCACAUCCUGCUUAUCUGCAACUG--ACUGCAGGAGAUAUGUGUGCGGCACAG ...-....(((((((-...))).))))..(((...((((((...(((-(((((((....)))))))))..((((((....--..)))))).)...))))))..)))... ( -32.40) >DroSec_CAF1 41267 104 + 1 AAU-GAUUUUCGAAU-CCAGUUACGAACAUGCAAUUACACGGGGCAG-CAGGAUGGGCACAUCCUGCUUAUCUGCAUCUG--ACUGCAGGAGAUAUGUGUGCGGCACAG ...-....(((((((-...))).))))..(((...((((((...(((-(((((((....)))))))))..((((((....--..)))))).)...))))))..)))... ( -32.20) >DroSim_CAF1 48243 104 + 1 AAU-GAUUUUCGAAU-CGAGUUACGAACAUGCAAUUACACGGGCCAG-CAGGAUGGGCACAUCCUGCUUAUCUGCAUCUG--ACUGCAGGAGAUAUGUGUGCGGCACAG ..(-((((....)))-))...........(((...((((((....((-(((((((....)))))))))..((((((....--..)))))).....))))))..)))... ( -32.10) >DroEre_CAF1 41989 80 + 1 AAU-CAUUUUUGAAC-CCAGUUGCAU------------ACGGCGUUG-CAGGAUGAGCACAUCCUGCUUAUCUGCAUCUG--ACUGCAG------------CCGCAUAG ..(-((....)))..-...((((((.------------.(((...((-(((.(((((((.....)))))))))))).)))--..)))))------------)....... ( -24.60) >DroYak_CAF1 43417 89 + 1 AAU-AAUGGUUGAAU-CCAGUUACACACAUGCAAUUACACGGGGCUG-CAGGAUGAGCACAUCCUGCUUAUCUGCAUCUG--GCUGCAG---------------GAUAG ...-.........((-((.((..(.((.(((((........((...(-(((((((....))))))))...))))))).))--)..)).)---------------))).. ( -23.70) >DroAna_CAF1 49388 104 + 1 AGUACAUUUUUGAAUUUCUGUUACGAAUAUGCAAUUACACAAGGAUGCCAGGAUGAGCACAUCCUGCUUAUCUGCAUCUCCUGCUGCAGGAAGCAUCAUCCUGG----- ....((((((((......(((.........)))......))))))))(((((((((((...((((((......(((.....))).)))))).)).)))))))))----- ( -32.60) >consensus AAU_GAUUUUCGAAU_CCAGUUACGAACAUGCAAUUACACGGGGCAG_CAGGAUGAGCACAUCCUGCUUAUCUGCAUCUG__ACUGCAGGAGAUAUGUGUGCGGCACAG ..............................................(.(((((((....))))))))....(((((........))))).................... (-15.55 = -15.55 + -0.00)
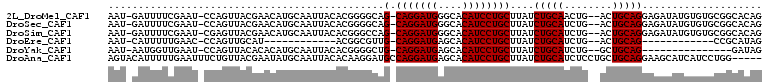

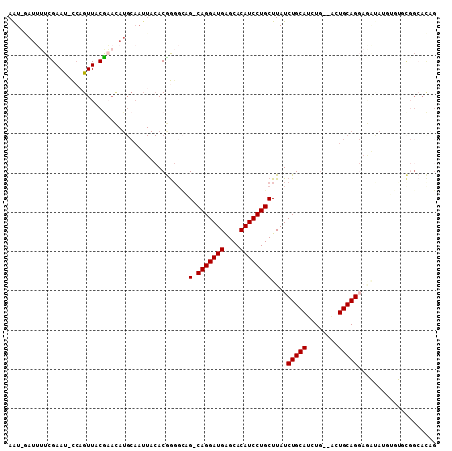
| Location | 10,958,308 – 10,958,412 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10958308 104 - 22407834 CUGUGCCGCACACAUAUCUCCUGCAGU--CAGUUGCAGAUAAGCAGGAUGUGCCCAUCCUG-CUGCCCCGUGUAAUUGCAUGUUCGUAACUGG-AUUCGAAAAUC-AUU .((((.....)))).............--(((((((.....(((((((((....)))))))-))....(((((....)))))...)))))))(-(((....))))-... ( -30.10) >DroSec_CAF1 41267 104 - 1 CUGUGCCGCACACAUAUCUCCUGCAGU--CAGAUGCAGAUAAGCAGGAUGUGCCCAUCCUG-CUGCCCCGUGUAAUUGCAUGUUCGUAACUGG-AUUCGAAAAUC-AUU ..((((...((((.......(((((..--....)))))...(((((((((....)))))))-)).....))))....)))).((((.......-...))))....-... ( -30.30) >DroSim_CAF1 48243 104 - 1 CUGUGCCGCACACAUAUCUCCUGCAGU--CAGAUGCAGAUAAGCAGGAUGUGCCCAUCCUG-CUGGCCCGUGUAAUUGCAUGUUCGUAACUCG-AUUCGAAAAUC-AUU ..((((...((((.......(((((..--....)))))...(((((((((....)))))))-)).....))))....)))).((((.......-...))))....-... ( -30.30) >DroEre_CAF1 41989 80 - 1 CUAUGCGG------------CUGCAGU--CAGAUGCAGAUAAGCAGGAUGUGCUCAUCCUG-CAACGCCGU------------AUGCAACUGG-GUUCAAAAAUG-AUU .(((((((------------(((((..--....)))).....((((((((....)))))))-)...)))))------------))).......-...........-... ( -29.70) >DroYak_CAF1 43417 89 - 1 CUAUC---------------CUGCAGC--CAGAUGCAGAUAAGCAGGAUGUGCUCAUCCUG-CAGCCCCGUGUAAUUGCAUGUGUGUAACUGG-AUUCAACCAUU-AUU .....---------------.((...(--(((.((((.(...((((((((....)))))))-).....(((((....)))))).)))).))))-...))......-... ( -27.20) >DroAna_CAF1 49388 104 - 1 -----CCAGGAUGAUGCUUCCUGCAGCAGGAGAUGCAGAUAAGCAGGAUGUGCUCAUCCUGGCAUCCUUGUGUAAUUGCAUAUUCGUAACAGAAAUUCAAAAAUGUACU -----(((((((((.((.((((((.(((.....)))......))))))...)))))))))))......(((((....)))))(((......)))............... ( -31.40) >consensus CUGUGCCGCACACAUAUCUCCUGCAGU__CAGAUGCAGAUAAGCAGGAUGUGCCCAUCCUG_CAGCCCCGUGUAAUUGCAUGUUCGUAACUGG_AUUCAAAAAUC_AUU ....................(((((........)))))....((((((((....))))))).).....(((((....)))))........................... (-17.41 = -17.77 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:26 2006