| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,905,391 – 10,905,500 |
| Length | 109 |
| Max. P | 0.841758 |

| Location | 10,905,391 – 10,905,500 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -15.23 |
| Energy contribution | -16.12 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
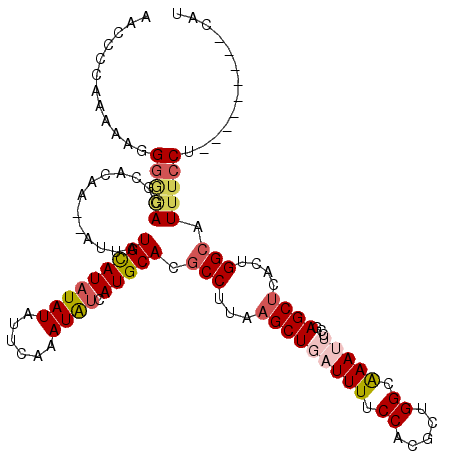
>2L_DroMel_CAF1 10905391 109 - 22407834 AGCCCCAAAAAGGGAGACGCACAA--AUUAUGCAUAUAUAUUCAAAUAUCAUGCACGCCUUAAGCUGAUUUUCCACGCUGGCAAAUUCCGAGCUCACUGGCAUUUCCU---------UAU ..........(((((((.......--....(((((((((......)))).))))).(((...(((((((((.((.....)).)))))...))))....))).))))))---------).. ( -23.60) >DroSec_CAF1 47173 109 - 1 AACCCCAAAAAGGGGGACGCACAA--AUUAUGCAUAUAUAUUCAAAUAUCAUGCACGCCUUAAGCUGAUUUUCCACGCUGGCAAAUUCCGAGCUCACUGGCAUUGCCU---------CAU ..((((.....))))((.(((...--....(((((((((......)))).))))).(((...(((((((((.((.....)).)))))...))))....)))..))).)---------).. ( -28.80) >DroSim_CAF1 46545 109 - 1 AACCCCAAAAAGGGGGACUCACAA--AUUAUGCAUAUAUAUUCAAAUAUCAUGCACGCCUUAAGCUGAUUUUCCACGCUGGCAAAUUCCGAGCUCACUGGCAUUUCCU---------CAU ..((((.....)))).........--....(((((((((......)))).))))).(((...(((((((((.((.....)).)))))...))))....))).......---------... ( -24.70) >DroWil_CAF1 64859 112 - 1 ------AAAAAAGAAAAAGGAAAAUUAUUAUGUAUAUAUAUUCAAAUGGCAUGCACGCCUAAAGCUGAUUUUCCACGCUGGGAAACGCUUAGCUU--UGGAAUUUACCACCCAACUCCAC ------............((.(((((....(((((.(((......)))..)))))...((((((((((((((((.....))))))...)))))))--)))))))).))............ ( -22.10) >DroYak_CAF1 46383 107 - 1 AACCC--AAAAGGGGGACGCACAA--AUUAUGCAUAUAUAUUCAAAUAUCAUGCACGCCUUAAGCUGAUUUUCCACGCUGGAAAAUUCCGAGCUCACUGGCAUUUCCU---------CAU .....--....((((((.......--....(((((((((......)))).))))).(((...((((((((((((.....))))))))...))))....)))..)))))---------).. ( -28.70) >DroAna_CAF1 49850 108 - 1 AAUUGUACU-ACGGGUAAGGUAAA--UGUAUGUAUAUAUAUUCAAAUAUCAUGCACGCCUUAAGCUGAUUUUCCACGCUGGUGAACGUCAAGCCUCCAGGCAUUUCCU---------CAU ..(((.((.-.(((.((((((...--((((((.((((........)))))))))).))))))..)))......(((....)))...)))))(((....))).......---------... ( -23.00) >consensus AACCCCAAAAAGGGGGACGCACAA__AUUAUGCAUAUAUAUUCAAAUAUCAUGCACGCCUUAAGCUGAUUUUCCACGCUGGCAAAUUCCGAGCUCACUGGCAUUUCCU_________CAU ............(((((.............(((((((((......)))).))))).(((...(((((((((.((.....)).)))))...))))....))).)))))............. (-15.23 = -16.12 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:14 2006