
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,899,095 – 10,899,212 |
| Length | 117 |
| Max. P | 0.879929 |

| Location | 10,899,095 – 10,899,212 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
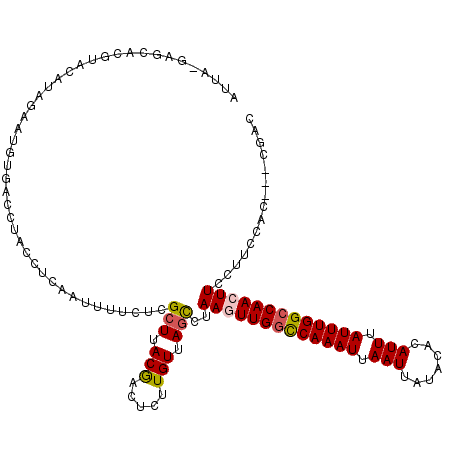
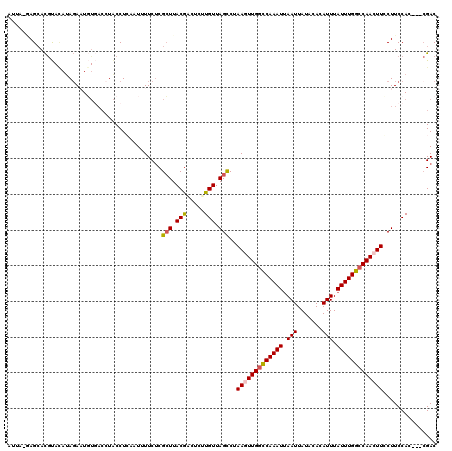
>2L_DroMel_CAF1 10899095 117 + 22407834 AUUAUAAGCACAUACAAAGAAUGUGACCUACCUCAAUUUUCUCGCUUACGACUCUUGUUAGCCUAAGUUGGCCAAAUUAAUUAUACACAUUUAUUUGGCCAACUUACUUCCAC---AGAC .(..(((((((((.......))))((......)).........)))))..).(((((..((..((((((((((((((.(((.......))).))))))))))))))))..)).---))). ( -26.60) >DroSec_CAF1 38717 116 + 1 AUUA-GAGCACGUAUAUAGAUUGUGACCUACCUGAAUAUUCUCGCUUACGCCUCUUGUUAGUCUAAGUUGACCAAAUUAAUUAUACACAUUUAUUUGGCCAACUUCCUUCCAG---CGAC ....-(((..((((...(((.(((...........))).)))....)))).)))(((((.(...((((((.((((((.(((.......))).)))))).))))))....).))---))). ( -19.30) >DroSim_CAF1 40037 116 + 1 AUUA-GAGAACGUACAUAGAUUGUGACCUACCUGAAUAUUCUCGCUUACGCCUCUUGUUAGCCUAAGUUGGCCAAAUUAAUUAUACACAUUUAUUUGGCCAACUUCCUUCCAG---CGAC ....-(((((..(.(((((........)))..)).)..))))).....(((....((..((...(((((((((((((.(((.......))).))))))))))))).))..)))---)).. ( -27.80) >DroEre_CAF1 38977 103 + 1 -----UAGCACGC--AUAGAAUU-----UGCCUCAAUUUUCUCGGUUACAA--CUUGUUAGCCCAAGUUGGUCAAAUUAAUUAUACACAUUUAUUUGGCCAAGUUGCUUCCAC---CGAC -----......((--(.......-----)))..........(((((.....--...(..(((..((.((((((((((.(((.......))).)))))))))).)))))..)))---))). ( -20.20) >DroYak_CAF1 39726 113 + 1 -----UAGCAUGCACAUAUAAUUUUACUUACCUCAAUUUUCCCGCUUACGU--UAUGUUAGCCAAAGUUGGUCAAAUGAAUUAUACACAUUUAUUUGGCCAAGUUCCUUCCACCGCCGAC -----..((..................................(((.((..--...)).)))..((.((((((((((((((.......)))))))))))))).)).........)).... ( -19.30) >consensus AUUA_GAGCACGUACAUAGAAUGUGACCUACCUCAAUUUUCUCGCUUACGACUCUUGUUAGCCUAAGUUGGCCAAAUUAAUUAUACACAUUUAUUUGGCCAACUUCCUUCCAC___CGAC ...........................................(((.(((.....))).)))..(((((((((((((.(((.......))).)))))))))))))............... (-15.00 = -15.24 + 0.24)

| Location | 10,899,095 – 10,899,212 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.60 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
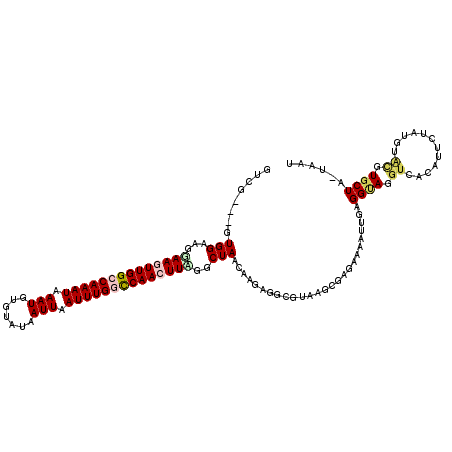
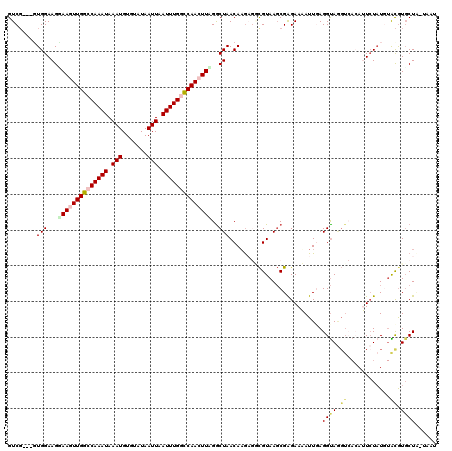
>2L_DroMel_CAF1 10899095 117 - 22407834 GUCU---GUGGAAGUAAGUUGGCCAAAUAAAUGUGUAUAAUUAAUUUGGCCAACUUAGGCUAACAAGAGUCGUAAGCGAGAAAAUUGAGGUAGGUCACAUUCUUUGUAUGUGCUUAUAAU .(((---.(((...((((((((((((((.(((.......))).))))))))))))))..)))...))).(((....)))..........(((((.(((((.......))))))))))... ( -34.50) >DroSec_CAF1 38717 116 - 1 GUCG---CUGGAAGGAAGUUGGCCAAAUAAAUGUGUAUAAUUAAUUUGGUCAACUUAGACUAACAAGAGGCGUAAGCGAGAAUAUUCAGGUAGGUCACAAUCUAUAUACGUGCUC-UAAU ....---.((..((.(((((((((((((.(((.......))).)))))))))))))...))..))(((((((((...(((....)))..((((((....)))))).))))).)))-)... ( -30.90) >DroSim_CAF1 40037 116 - 1 GUCG---CUGGAAGGAAGUUGGCCAAAUAAAUGUGUAUAAUUAAUUUGGCCAACUUAGGCUAACAAGAGGCGUAAGCGAGAAUAUUCAGGUAGGUCACAAUCUAUGUACGUUCUC-UAAU .((.---.((..((.(((((((((((((.(((.......))).)))))))))))))...))..)).)).((....))((((((....(.((((((....)))))).)..))))))-.... ( -35.00) >DroEre_CAF1 38977 103 - 1 GUCG---GUGGAAGCAACUUGGCCAAAUAAAUGUGUAUAAUUAAUUUGACCAACUUGGGCUAACAAG--UUGUAACCGAGAAAAUUGAGGCA-----AAUUCUAU--GCGUGCUA----- .(((---((....((((((((..(((((.(((.......))).))))).((.....)).....))))--)))).))))).........((((-----..(.....--)..)))).----- ( -22.70) >DroYak_CAF1 39726 113 - 1 GUCGGCGGUGGAAGGAACUUGGCCAAAUAAAUGUGUAUAAUUCAUUUGACCAACUUUGGCUAACAUA--ACGUAAGCGGGAAAAUUGAGGUAAGUAAAAUUAUAUGUGCAUGCUA----- ...((((((.((......)).)))......((((((((((((.(((((.((((.(((.(((.((...--..)).))).)))...))).).)))))..))))))))..))))))).----- ( -20.20) >consensus GUCG___GUGGAAGGAAGUUGGCCAAAUAAAUGUGUAUAAUUAAUUUGGCCAACUUAGGCUAACAAGAGGCGUAAGCGAGAAAAUUGAGGUAGGUCACAUUCUAUGUACGUGCUA_UAAU ........(((...((((((((((((((.(((.......))).))))))))))))))..)))..........................((((.((............)).))))...... (-15.88 = -16.92 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:09 2006