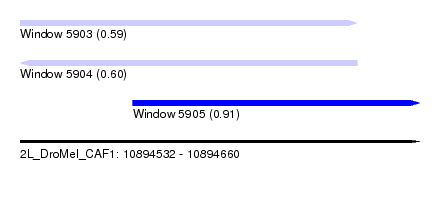
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,894,532 – 10,894,660 |
| Length | 128 |
| Max. P | 0.909598 |

| Location | 10,894,532 – 10,894,640 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -21.49 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10894532 108 + 22407834 UUGCCAGUCAGUCGACAAGGC---AAGUUUAUUGUACUU--UGUACUU--GGUGGUU----AUGGCAUGAUGUAUUCGUGCCAUGGCAUGUAAUUCAUCAGCCGGCG-AGAUGAAUGAGU (((((.(((....)))..(((---.........((((..--.)))).(--(((((((----(((((((((.....))))))))))))........))))))))))))-)........... ( -36.20) >DroSec_CAF1 34083 104 + 1 UUGCCA----GUCGACAAGGC---AAGUUUAUUGUACUU--UGUACUU--GGGGGUU----AUGGCAUGAUGUAUUCGUGCCACGGCAUGUAAUUCAUCAGCCGGCG-AGAUGAAUGAGU ((((((----..(..((((((---((((.......)).)--))).)))--)..)..)----..(((.(((((.((((((((....))))).))).))))))))))))-)........... ( -33.80) >DroSim_CAF1 34972 104 + 1 UUGCCA----GUCGACAAGGC---AAGUUUAUUGUACUU--UGUACUU--GGGGGUU----AUGGCAUGAUGUAUUCGUGCCACGGCAUGUAAUUCAUCAGCCGGCG-AGAUGAAUGAGU ((((((----..(..((((((---((((.......)).)--))).)))--)..)..)----..(((.(((((.((((((((....))))).))).))))))))))))-)........... ( -33.80) >DroEre_CAF1 34280 103 + 1 GU---UGCCAGUCGACAAGGC---GAGUUUAUUGUACUG--UGUACUU---GGGGUU----AUGGCAAGAUGUAUUCGUGCCAGGGCAUGUAAUUUAUCA-CCGGGG-AGAUGAAUGAGU .(---(((((..(..((((((---((((.......))).--))).)))---)..)..----.))))))........(((((....)))))..(((((((.-(....)-.))))))).... ( -26.90) >DroYak_CAF1 35050 110 + 1 UUGCCAGCCAGCCGACAAGGC---AAGUUUAUUGUGCUU--UGUACUUUGGGGGGUU----AUGGCAUGAUGUAUUCGUGCCAGGGCAUGUAAUUCAUCAGCCAUGG-AGAUGAAUGAGU ..(((..((((...(((((((---(.........)))))--)))...))))..)))(----(((((.(((((.((((((((....))))).))).))))))))))).-............ ( -39.10) >DroAna_CAF1 37553 113 + 1 UUGCCA-----CCGAGCAUGCAUGAAGUUUAUUGUCCUUUUUGUACAC--GGAAGACACAUGGGGUAUGAUGUAUUCGUGUCAGGGCAUGUAAUUCAUCUCGUGUCGCAAAUGAAUGAGC ((((.(-----(((((.....(((((......((((((....(.((((--(((..(((((((...)))).))).)))))))))))))).....))))))))).)).)))).......... ( -28.80) >consensus UUGCCA____GUCGACAAGGC___AAGUUUAUUGUACUU__UGUACUU__GGGGGUU____AUGGCAUGAUGUAUUCGUGCCAGGGCAUGUAAUUCAUCAGCCGGCG_AGAUGAAUGAGU ..........................(((((((((((.....))))................((((.(((((.((((((((....))))).))).))))))))).....))))))).... (-21.49 = -21.88 + 0.39)

| Location | 10,894,532 – 10,894,640 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -14.72 |
| Energy contribution | -16.08 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10894532 108 - 22407834 ACUCAUUCAUCU-CGCCGGCUGAUGAAUUACAUGCCAUGGCACGAAUACAUCAUGCCAU----AACCACC--AAGUACA--AAGUACAAUAAACUU---GCCUUGUCGACUGACUGGCAA ............-....((((((((.(((.(.(((....))).)))).))))).)))..----.......--..((((.--..))))........(---(((..(((....))).)))). ( -26.00) >DroSec_CAF1 34083 104 - 1 ACUCAUUCAUCU-CGCCGGCUGAUGAAUUACAUGCCGUGGCACGAAUACAUCAUGCCAU----AACCCCC--AAGUACA--AAGUACAAUAAACUU---GCCUUGUCGAC----UGGCAA ............-.(((((((((((.(((.(.(((....))).)))).))))).)))..----...(..(--(((....--((((.......))))---..))))..)..----.))).. ( -25.10) >DroSim_CAF1 34972 104 - 1 ACUCAUUCAUCU-CGCCGGCUGAUGAAUUACAUGCCGUGGCACGAAUACAUCAUGCCAU----AACCCCC--AAGUACA--AAGUACAAUAAACUU---GCCUUGUCGAC----UGGCAA ............-.(((((((((((.(((.(.(((....))).)))).))))).)))..----...(..(--(((....--((((.......))))---..))))..)..----.))).. ( -25.10) >DroEre_CAF1 34280 103 - 1 ACUCAUUCAUCU-CCCCGG-UGAUAAAUUACAUGCCCUGGCACGAAUACAUCUUGCCAU----AACCCC---AAGUACA--CAGUACAAUAAACUC---GCCUUGUCGACUGGCA---AC ............-..((((-((((((...........(((((.((.....)).))))).----......---..((((.--..)))).........---...))))).)))))..---.. ( -18.50) >DroYak_CAF1 35050 110 - 1 ACUCAUUCAUCU-CCAUGGCUGAUGAAUUACAUGCCCUGGCACGAAUACAUCAUGCCAU----AACCCCCCAAAGUACA--AAGCACAAUAAACUU---GCCUUGUCGGCUGGCUGGCAA ............-..((((((((((.(((.(.(((....))).)))).))))).)))))----..((..(((..(.(((--(.(((.........)---)).)))))...)))..))... ( -26.10) >DroAna_CAF1 37553 113 - 1 GCUCAUUCAUUUGCGACACGAGAUGAAUUACAUGCCCUGACACGAAUACAUCAUACCCCAUGUGUCUUCC--GUGUACAAAAAGGACAAUAAACUUCAUGCAUGCUCGG-----UGGCAA ..........((((.((.(((((((((.....((.(((.(((((((.(((.(((.....)))))).)).)--))))......))).))......))))).....)))))-----).)))) ( -23.70) >consensus ACUCAUUCAUCU_CGCCGGCUGAUGAAUUACAUGCCCUGGCACGAAUACAUCAUGCCAU____AACCCCC__AAGUACA__AAGUACAAUAAACUU___GCCUUGUCGAC____UGGCAA ....(((((((..........)))))))....((((.(((((.((.....)).)))))................((((.....))))............................)))). (-14.72 = -16.08 + 1.36)


| Location | 10,894,568 – 10,894,660 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -19.06 |
| Energy contribution | -18.87 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10894568 92 + 22407834 -UGUACUU--GGUGGUU----AUGGCAUGAUGUAUUCGUGCCAUGGCAUGUAAUUCAUCAGCCGGCG-AGAUGAAUGAGUCGUAUUCAUAUA--------------UAU------AUUCA -....(((--(.(((((----(((((((((.....))))))))).(.(((.....))))))))).))-))(((((((.....)))))))...--------------...------..... ( -27.50) >DroSec_CAF1 34115 90 + 1 -UGUACUU--GGGGGUU----AUGGCAUGAUGUAUUCGUGCCACGGCAUGUAAUUCAUCAGCCGGCG-AGAUGAAUGAGUCGUAUUCAUAUA----------------U------AUUCA -....(((--(......----.((((((((.....))))))))((((.((........)))))).))-))(((((((.....)))))))...----------------.------..... ( -26.30) >DroSim_CAF1 35004 106 + 1 -UGUACUU--GGGGGUU----AUGGCAUGAUGUAUUCGUGCCACGGCAUGUAAUUCAUCAGCCGGCG-AGAUGAAUGAGUCGUAUUCAUAUAUAUUCACAUUCAUAUAU------AUUCA -....(((--(......----.((((((((.....))))))))((((.((........)))))).))-))(((((((.(..((((......)))).).)))))))....------..... ( -29.00) >DroEre_CAF1 34313 100 + 1 -UGUACUU---GGGGUU----AUGGCAAGAUGUAUUCGUGCCAGGGCAUGUAAUUUAUCA-CCGGGG-AGAUGAAUGAGUCGUAUUCAUA--UA--CACAUACUCGUAU------AUUCA -(((((..---(((((.----.(((((.((.....)).)))))..))((((.(((((((.-(....)-.))))))).....((((....)--))--))))).)))))))------).... ( -24.20) >DroYak_CAF1 35086 112 + 1 -UGUACUUUGGGGGGUU----AUGGCAUGAUGUAUUCGUGCCAGGGCAUGUAAUUCAUCAGCCAUGG-AGAUGAAUGAGUCGUAUUCAUAUAUA--CACAUACGAGUAUACUCGUAUUCA -((((..........((----(((((.(((((.((((((((....))))).))).))))))))))))-..(((((((.....)))))))...))--)).(((((((....)))))))... ( -35.90) >DroAna_CAF1 37588 101 + 1 UUGUACAC--GGAAGACACAUGGGGUAUGAUGUAUUCGUGUCAGGGCAUGUAAUUCAUCUCGUGUCGCAAAUGAAUGAGCCGUACACAUUUAUU--CA---------GU------AUUCA ((((((((--(((.((.(((((.(..((((.....))))..)....)))))...)).)).))))).)))).((((((((..(....).))))))--))---------..------..... ( -23.00) >consensus _UGUACUU__GGGGGUU____AUGGCAUGAUGUAUUCGUGCCAGGGCAUGUAAUUCAUCAGCCGGCG_AGAUGAAUGAGUCGUAUUCAUAUAUA__CA________UAU______AUUCA ..((((................((((((((.....)))))))).(((.....(((((((..........)))))))..)))))))................................... (-19.06 = -18.87 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:07 2006