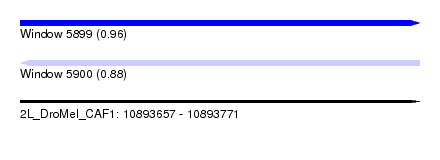
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,893,657 – 10,893,771 |
| Length | 114 |
| Max. P | 0.959689 |

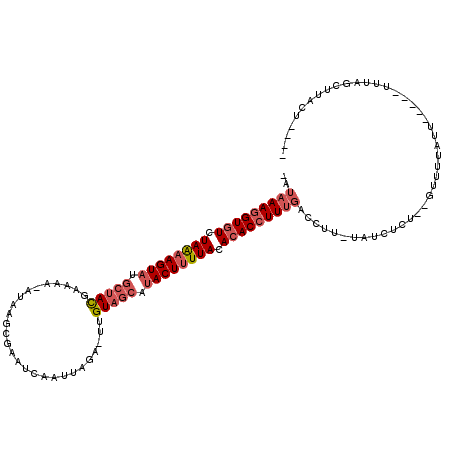
| Location | 10,893,657 – 10,893,771 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.09 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -13.87 |
| Energy contribution | -16.10 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10893657 114 + 22407834 ----AGAAAGCUAAAAACAAAAAUAUACCAAGAGAUAUAAGAUCAAAGGUGUGUAAAAGUAUGAAACUAAUUCACUUGAAUCGUUUAU-UUUUCGUAGCUUACUAUUAGAUACAUUUAU- ----((.((((((.........(((((((....(((.....)))...)))))))(((((((...(((..((((....)))).))))))-))))..)))))).))...............- ( -15.50) >DroEre_CAF1 33453 109 + 1 AACUCCUAAGCUAAA-----ACUAAAAC--CGGGAUA-AAGGUCAAAGGUGUGUAAAAGUGUUCUACA---CUAAUGGAUUCGCUUAUUUUUUCAUAGCAUACUUCUAGACACCUUUAUC ...............-----......((--(......-..))).((((((((.((.((((((((((..---....))))...(((...........))))))))).)).))))))))... ( -21.30) >DroYak_CAF1 34228 105 + 1 ----AGUAGGCUAAAU----AAUAAAAC--AGAGGUG-CAGCACAAAGGUGUGUAAAAGUAGGCUACAAUUCUAAUAGACUCGCUUUUACUUUCGUAGCAUACUUUUAAACACCUU---- ----.....(((..((----........--....)).-.)))...(((((((.((((((((.(((((.....(((.((.....)).))).....))))).)))))))).)))))))---- ( -24.50) >consensus ____AGUAAGCUAAA_____AAUAAAAC__AGAGAUA_AAGAUCAAAGGUGUGUAAAAGUAUGCUACAA_UCUAAUAGAAUCGCUUAU_UUUUCGUAGCAUACUUUUAGACACCUUUAU_ ............................................((((((((.((((((((.(((((...........................))))).)))))))).))))))))... (-13.87 = -16.10 + 2.23)

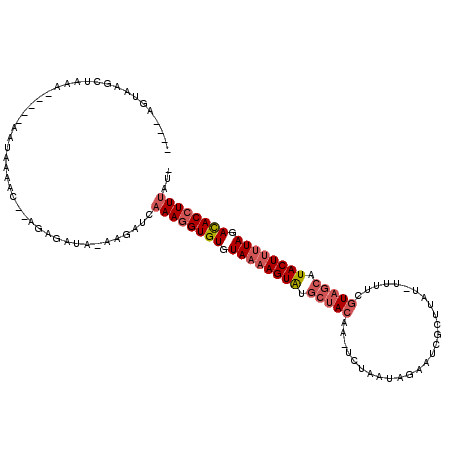
| Location | 10,893,657 – 10,893,771 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.09 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -13.20 |
| Energy contribution | -15.76 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10893657 114 - 22407834 -AUAAAUGUAUCUAAUAGUAAGCUACGAAAA-AUAAACGAUUCAAGUGAAUUAGUUUCAUACUUUUACACACCUUUGAUCUUAUAUCUCUUGGUAUAUUUUUGUUUUUAGCUUUCU---- -...............((.((((((...(((-(((((.(((.((((((.(..(((.....)))..).)))....)))))).((((((....))))))..)))))))))))))).))---- ( -16.50) >DroEre_CAF1 33453 109 - 1 GAUAAAGGUGUCUAGAAGUAUGCUAUGAAAAAAUAAGCGAAUCCAUUAG---UGUAGAACACUUUUACACACCUUUGACCUU-UAUCCCG--GUUUUAGU-----UUUAGCUUAGGAGUU ..(((((((((.(((((((.((((((......)).)))).........(---(.....))))))))).))))))))).....-..(((.(--(((.....-----...))))..)))... ( -23.30) >DroYak_CAF1 34228 105 - 1 ----AAGGUGUUUAAAAGUAUGCUACGAAAGUAAAAGCGAGUCUAUUAGAAUUGUAGCCUACUUUUACACACCUUUGUGCUG-CACCUCU--GUUUUAUU----AUUUAGCCUACU---- ----(((((((.((((((((.(((((....)))...((((.((.....)).)))).)).)))))))).)))))))...((((-.......--........----...)))).....---- ( -25.77) >consensus _AUAAAGGUGUCUAAAAGUAUGCUACGAAAA_AUAAGCGAAUCAAUUAGA_UUGUAGCAUACUUUUACACACCUUUGACCUU_UAUCUCU__GUUUUAUU_____UUUAGCUUACU____ ..(((((((((.((((((((.(((((...........................))))).)))))))).)))))))))........................................... (-13.20 = -15.76 + 2.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:02 2006