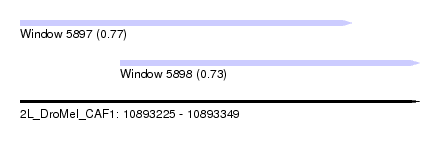
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,893,225 – 10,893,349 |
| Length | 124 |
| Max. P | 0.773429 |

| Location | 10,893,225 – 10,893,328 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 89.32 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.54 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10893225 103 + 22407834 -UCCCCAUUGGCCCCCCAAAAAAAAGUGGUAGCAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACCCAAGUGCC -......((((....))))......(..(((((..((((((....(((....)))...........))))))..)))))..).(..((((......))))..). ( -23.66) >DroSec_CAF1 32812 98 + 1 -CCCCCAUUGGCCC-----CCAAAAAUGGUAGCAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACCCAAGUGCC -......((((...-----))))..((((((((..((((((....(((....)))...........))))))..)))))))).(..((((......))))..). ( -22.96) >DroSim_CAF1 33678 103 + 1 -CCCCCAUUGGCCCUCCCCCAAAAAAUGGUAGCAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACCCAAGUGCC -......((((.......))))...((((((((..((((((....(((....)))...........))))))..)))))))).(..((((......))))..). ( -23.46) >DroEre_CAF1 33014 97 + 1 ---CCCACUGGCCCC----CAAAAAAUGGUAGCAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACCCAAGUGCC ---.......((..(----((.....)))..))...........((((..(((((...............)))))..))))..(..((((......))))..). ( -22.06) >DroYak_CAF1 33765 100 + 1 GCCCCCAUUGGCCCC----CAAAAAAUGGUAGCAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACCCAAGUGCC (((......)))...----......((((((((..((((((....(((....)))...........))))))..)))))))).(..((((......))))..). ( -24.66) >DroAna_CAF1 36190 99 + 1 -AAAAAACAAGAACA----AAAAAAAUGGCUCCUACCGUUAUUGCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACCCAAGUACC -..............----.....(((((......)))))...(((((..(((((...............)))))..))))).(((((((......))))))). ( -22.26) >consensus _CCCCCAUUGGCCCC____CAAAAAAUGGUAGCAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACCCAAGUGCC .........................(..(((((..((((((....(((....)))...........))))))..)))))..).(((((((......))))))). (-20.62 = -20.54 + -0.08)

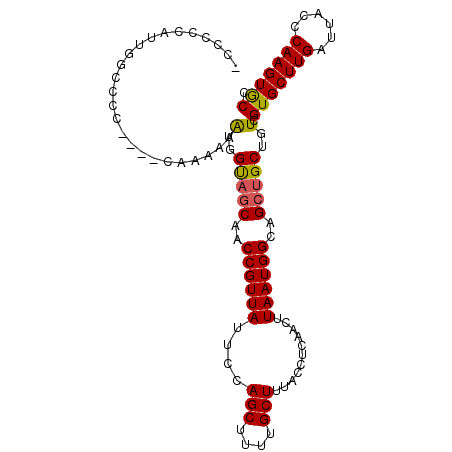
| Location | 10,893,256 – 10,893,349 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -19.37 |
| Consensus MFE | -14.44 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10893256 93 + 22407834 -------CAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACC-CAAGUGCCACGCCUCUU------------CCGCC----CAUCCA---C -------....(((.....((((..(((((...............)))))..))))..(..((((......-))))..).)))......------------.....----......---. ( -17.46) >DroSim_CAF1 33709 93 + 1 -------CAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACC-CAAGUGCCACGCCUCUU------------CCGCC----CAUCCA---C -------....(((.....((((..(((((...............)))))..))))..(..((((......-))))..).)))......------------.....----......---. ( -17.46) >DroEre_CAF1 33039 97 + 1 -------CAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACC-CAAGUGCCACGCCCCAU------------CCGCCGCCCCUUCCG---C -------....(((.....((((..(((((...............)))))..))))..(..((((......-))))..).)))......------------...............---. ( -17.46) >DroWil_CAF1 46764 104 + 1 UUACGUUACUCCGUUACUCCAUCCAAGGCUUUAACUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACCCCGAGUACCACGCCUUCUUUC---------CUGUC----CUUUCC---U ........................(((((................(((((...)))))(((((((.......)))))))...))))).....---------.....----......---. ( -17.90) >DroYak_CAF1 33793 95 + 1 -------CAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACC-CAAGUGCCACGCCCCUU------------CCGCAGCCCCAUCC----- -------.............(((....))).............((((..((((((((.(..((((......-))))..).)))......------------..))))).))))..----- ( -20.40) >DroAna_CAF1 36217 110 + 1 -------CUACCGUUAUUGCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACC-CAAGUACCACGCCUCCUUUUAGGUGCCUCCCGCCUU--CGUCCUCGCC -------...........((((((...(((...............))))))))).((.(((((((......-))))))).))((........(((((.....))))).--.......)). ( -25.55) >consensus _______CAACCGUUAUUCCAGCUUUUGCUUUACCUCAACUUAAUGGCAGCUGCUGUCGUGCUUGAUUACC_CAAGUGCCACGCCUCUU____________CCGCC____CAUCCA___C ...........(((.....((((..(((((...............)))))..))))..(((((((.......))))))).)))..................................... (-14.44 = -14.58 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:00:00 2006