
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,892,460 – 10,892,562 |
| Length | 102 |
| Max. P | 0.806555 |

| Location | 10,892,460 – 10,892,562 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.65 |
| Mean single sequence MFE | -23.06 |
| Consensus MFE | -7.96 |
| Energy contribution | -9.60 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10892460 102 + 22407834 ----------CU---AACCAAAGUUCGGAUCUCCAUAAACUUG-GAGCUUAGGCCCAUCAACUCUGCGUAUA---UGGCCAAAAACGCCAUAGCCC-CGCCUCCGCUUUUUUGGCCAACA ----------..---..((((((..((((.(((((......))-)))..................(((..((---((((.......))))))....-))).))))...))))))...... ( -26.40) >DroSec_CAF1 32058 104 + 1 ----------CUGAAAACCAAAGUUCGGAUCUCCAUAAACCUG-GAGCUUAGGCCCAUCAACUCUGCGUAUA---UGGCCAAAAACGCCAUUGCC--CGCCCCCCGCUUUUUGGCCAACA ----------.......((((((..(((..(((((......))-)))..................(((...(---((((.......)))))....--)))...)))..))))))...... ( -24.50) >DroSim_CAF1 32930 115 + 1 AAACUGAAAACUGAAAACCAAAGUUCGGAUCUCCAUAAACUUG-GAGCUUAGGCCCAUCAACUCUGCGUAUA---UGGCCAAAAACGCCAUUGCCC-CGCCCCCCGCUUUUUGGCCAACA ...((((...(((((........)))))..(((((......))-))).))))....................---((((((((((.((....((..-.)).....))))))))))))... ( -26.10) >DroEre_CAF1 32194 103 + 1 ----------CU---AACCAAAGUUCGGAUCUCCAAAAGCUUG-GAGCUUAGGCCCAUCAAUUUUGGCCAUA---UGACCAGAAACCCCAUGCCCCAUGCCCCACCCUUUUUGGCCAACA ----------..---.......(((.((....(((((((..((-(.((...(((.......((((((.(...---.).)))))).......)))....)).)))...)))))))))))). ( -21.64) >DroYak_CAF1 33004 79 + 1 ----------CU---AACCAAAGUUCGGAUCUCCAUAAGCUCC-AAGCUUAGGCCCAUCAAUUCUGGCCAU---------------------------CGCCCACCCUUUUUGGCCACCA ----------..---..((.(((((.(((.((.....)).)))-.))))).))...........((((((.---------------------------.............))))))... ( -17.94) >DroAna_CAF1 35108 92 + 1 ----------CU---AACUAAAGUUU--------UUGAACGUGCGAGCUUAGCCACAUCAAUUUGGCUCAAACACUGGGCCAUCUCGCCAAAGCCC-C------AGAGCUCGGUCCGCCA ----------..---...........--------......(((((((((((((((........)))))........((((............))))-.------.)))))))...))).. ( -21.80) >consensus __________CU___AACCAAAGUUCGGAUCUCCAUAAACUUG_GAGCUUAGGCCCAUCAACUCUGCGCAUA___UGGCCAAAAACGCCAUAGCCC_CGCCCCCCCCUUUUUGGCCAACA .................((.((((((((....))..........)))))).))......................((((((((((......................))))))))))... ( -7.96 = -9.60 + 1.64)

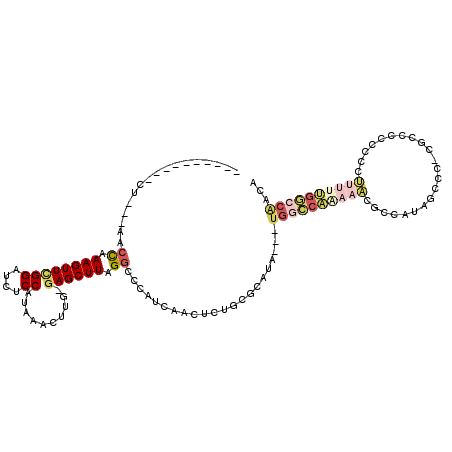
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:59 2006