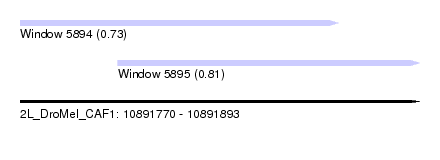
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,891,770 – 10,891,893 |
| Length | 123 |
| Max. P | 0.807037 |

| Location | 10,891,770 – 10,891,868 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.64 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -21.73 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
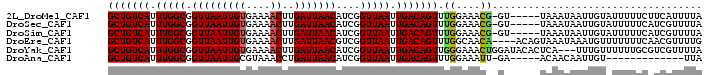
>2L_DroMel_CAF1 10891770 98 + 22407834 ----------GGCUGAUUUACCAAUUGAAAGUUCAUUUCAGCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUUGGAAACG-GU-----UA ----------(((((.(((.((((.(((((.....)))))(((((((.(((((.(((((((((....))..)))))))....))))).))))))))))))))))-))-----). ( -28.50) >DroSec_CAF1 31343 108 + 1 GAUUUCGGGUGGCUGAUUUACCAAUUGAAAGUUCAUUUCAGCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUUGGAAACG-GU-----UA .........((((((.(((.((((.(((((.....)))))(((((((.(((((.(((((((((....))..)))))))....))))).))))))))))))))))-))-----)) ( -29.00) >DroSim_CAF1 31740 108 + 1 GAUUUCGGGUGGCUGAUUUACCAAUUGAAAGUUCAUUUCAGCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUUGGAAACG-GU-----UA .........((((((.(((.((((.(((((.....)))))(((((((.(((((.(((((((((....))..)))))))....))))).))))))))))))))))-))-----)) ( -29.00) >DroEre_CAF1 31489 109 + 1 GAAUUCGGGUGGCUGA-UUACCAAUUGAAAGUUCAUUUCAGCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACGUCGGUUAAUUGACAGUUUGGCAACA----ACAGUA ...........((((.-........(((((.....)))))(((((((.((((((.((((((((....))..)))))))))))).....)))))))(((....))----))))). ( -26.40) >DroYak_CAF1 32274 102 + 1 ------------AUGAUUUACCAAUCGAAAGUUCAUUUCAGCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUGGGAAACUGGAUACACUC ------------........(((..(....)....(..(((((((((.(((((.(((((((((....))..)))))))....))))).)))))))))..)...)))........ ( -28.70) >DroAna_CAF1 34494 108 + 1 GAGUGAAGCCUGUUGAUAUACCGAUUGAAAAUUUAUUUCAGCUGUCAUUUGGCGGUUAAUUGCGUAAACCUGAUUAACAUCGGUUAAUUGACAGUUUGGAAAUU-GA-----AC ...(.((((.(((..((..(((((((((((.....)))))((((.....)))).((((((((.(....).))))))))))))))..))..))))))).).....-..-----.. ( -23.30) >consensus GA_UUCGGGUGGCUGAUUUACCAAUUGAAAGUUCAUUUCAGCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUUGGAAACG_GU_____UA ....................((((.(((((.....)))))(((((((.(((((.((((((((........))))))))....))))).)))))))))))............... (-21.73 = -21.95 + 0.22)

| Location | 10,891,800 – 10,891,893 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.21 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -17.49 |
| Energy contribution | -17.22 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10891800 93 + 22407834 GCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUUGGAAACG-GU-----UAAAUAAUUGUAUUUUUCUUCAUUUUA (((((((.(((((.(((((((((....))..)))))))....))))).)))))))..(....)(-(.-----.(((((....)))))..))........ ( -20.50) >DroSec_CAF1 31383 93 + 1 GCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUUGGAAACG-GU-----UAAAUAAUUGUAUUUUUCAUCGUUUUA (((((((.(((((.(((((((((....))..)))))))....))))).)))))))...((((((-((-----.(((((....)))))...)))))))). ( -25.10) >DroSim_CAF1 31780 93 + 1 GCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUUGGAAACG-GU-----UAAAUAAUUGUAUUUUUCAUCGUUUUA (((((((.(((((.(((((((((....))..)))))))....))))).)))))))...((((((-((-----.(((((....)))))...)))))))). ( -25.10) >DroEre_CAF1 31528 95 + 1 GCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACGUCGGUUAAUUGACAGUUUGGCAACA----ACAGUAAAUAAAUGUUUUUUUCAACGUUUUG (((((((.((((((.((((((((....))..)))))))))))).....)))))))(((....))----)........(((((((......))))))).. ( -22.10) >DroYak_CAF1 32302 96 + 1 GCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUGGGAAACUGGAUACACUCA---UUUGUUUUUUGCGUCGUUUUA (((((((.(((((.(((((((((....))..)))))))....))))).)))))))(..(((((..((.....)).---...)))))..).......... ( -21.40) >DroAna_CAF1 34534 80 + 1 GCUGUCAUUUGGCGGUUAAUUGCGUAAACCUGAUUAACAUCGGUUAAUUGACAGUUUGGAAAUU-GA-----ACAACAAUUGU-------------UUA (((((((.(((((.((((((((.(....).))))))))....))))).)))))))........(-((-----(((.....)))-------------))) ( -19.00) >consensus GCUGUCAUUUGGCGGUUAAUUGUGAAAACUUGAUUAACAUCGGUUAAUUGACAGUUUGGAAACG_GU_____UAAAUAAUUGUAUUUUUCAUCGUUUUA (((((((.(((((.((((((((........))))))))....))))).))))))).((....))................................... (-17.49 = -17.22 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:58 2006