| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,887,298 – 10,887,414 |
| Length | 116 |
| Max. P | 0.993808 |

| Location | 10,887,298 – 10,887,414 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -34.49 |
| Energy contribution | -35.32 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
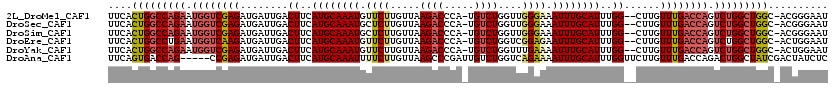
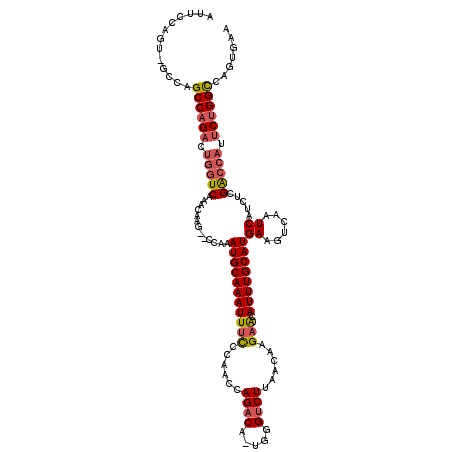
>2L_DroMel_CAF1 10887298 116 + 22407834 UUCACUGGCCAGAAUGGUCGAGAUGAUUGACUUCAUGCAAAUGUUCUUGUUAAGACCCA-UGUCUGGUUGGGAAAUUUGCAUUUGG--CUUGUUUGACCAGUCUGGCUGGC-ACGGGAAU ....(..((((((.((((((((......(.((..((((((((.((((((...((((...-.))))...))))))))))))))..))--)...)))))))).))))))..).-........ ( -41.60) >DroSec_CAF1 26951 116 + 1 UUCACUGGCCAGAAUGGUCGAGAUGAUUGACUUCAUGCAAAUGCUCUUGUUAAGACCCA-UGUCUGGUUGGGAAAUUUGCAUUUGG--CUUGUUUGACCAGUCUGGCUGGC-ACGGGAAU ....(..((((((.((((((((......(.((..((((((((..(((((...((((...-.))))...))))).))))))))..))--)...)))))))).))))))..).-........ ( -41.50) >DroSim_CAF1 27326 116 + 1 UUCACUGGCCAGAAUGGUCGAGAUGAUUGACUUCAUGCAAAUGCUCUUGUUAAGACCCA-UGUCUGGUUGGGAAAUUUGCAUUUGG--CUUGUUUGACCAGUCUGGCUGGC-ACGGGAAU ....(..((((((.((((((((......(.((..((((((((..(((((...((((...-.))))...))))).))))))))..))--)...)))))))).))))))..).-........ ( -41.50) >DroEre_CAF1 27265 116 + 1 UUCACUGGCCUGAAUGGUCAAGAUGAUUGACUUCAUGCAAAUGUUCUUGUUAAGACCCA-UGUCUGGUCGGAGAAUUUGCAUUUGG--CUUGUUUGACCAGUCUGGCUGGC-ACUGGAAU (((((..(((.((.((((((((......(.((..((((((((.((((.....((((...-.))))....)))).))))))))..))--)...)))))))).)).)))..).-..)))).. ( -37.00) >DroYak_CAF1 27829 116 + 1 UUCACUGGCCAGAAUGGUCGAGAUGAUUGACUUCAUGCAAAUGUUCUUGUUAAGACCCA-UGUCUGGUUUGAAAAUUUGCAUUUGG--CUUGUUUGACCAGUCUGGCUGGC-ACUGGAAU (((((..((((((.((((((((......(.((..((((((((.(((......((((...-.)))).....))).))))))))..))--)...)))))))).))))))..).-..)))).. ( -40.70) >DroAna_CAF1 28970 115 + 1 UUCAGUGACCAG-----CCGAGAUGAUUGACUUCAUGCAAAUUUUCUUGUUAAGCCCGAUUGUCUGGUCAGAAAAUUUGCAUUUGGUUCUUGUUUGACCAGACUGGCUAUCGACUAUCUC ............-----..((((((.((((......((((......))))..((((.....(((((((((((......((.....)).....))))))))))).)))).)))).)))))) ( -31.10) >consensus UUCACUGGCCAGAAUGGUCGAGAUGAUUGACUUCAUGCAAAUGUUCUUGUUAAGACCCA_UGUCUGGUUGGAAAAUUUGCAUUUGG__CUUGUUUGACCAGUCUGGCUGGC_ACGGGAAU ....(((((((((.((((((((........((..((((((((.((((.....((((.....))))....)))).))))))))..))......)))))))).))))))))).......... (-34.49 = -35.32 + 0.84)
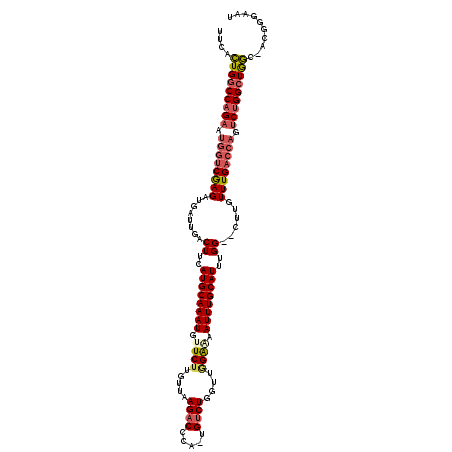
| Location | 10,887,298 – 10,887,414 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.17 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10887298 116 - 22407834 AUUCCCGU-GCCAGCCAGACUGGUCAAACAAG--CCAAAUGCAAAUUUCCCAACCAGACA-UGGGUCUUAACAAGAACAUUUGCAUGAAGUCAAUCAUCUCGACCAUUCUGGCCAGUGAA ........-((..((((((.(((((.......--....(((((((((((......((((.-...))))......))).))))))))((......)).....))))).))))))..))... ( -31.40) >DroSec_CAF1 26951 116 - 1 AUUCCCGU-GCCAGCCAGACUGGUCAAACAAG--CCAAAUGCAAAUUUCCCAACCAGACA-UGGGUCUUAACAAGAGCAUUUGCAUGAAGUCAAUCAUCUCGACCAUUCUGGCCAGUGAA ........-((..((((((.(((((......(--(.((((((......((((........-))))(((.....)))))))))))((((......))))...))))).))))))..))... ( -34.30) >DroSim_CAF1 27326 116 - 1 AUUCCCGU-GCCAGCCAGACUGGUCAAACAAG--CCAAAUGCAAAUUUCCCAACCAGACA-UGGGUCUUAACAAGAGCAUUUGCAUGAAGUCAAUCAUCUCGACCAUUCUGGCCAGUGAA ........-((..((((((.(((((......(--(.((((((......((((........-))))(((.....)))))))))))((((......))))...))))).))))))..))... ( -34.30) >DroEre_CAF1 27265 116 - 1 AUUCCAGU-GCCAGCCAGACUGGUCAAACAAG--CCAAAUGCAAAUUCUCCGACCAGACA-UGGGUCUUAACAAGAACAUUUGCAUGAAGUCAAUCAUCUUGACCAUUCAGGCCAGUGAA ....((.(-(...(((.((.(((((((.....--....(((((((((((..((((.....-..))))......)))..))))))))((......))...))))))).)).))))).)).. ( -28.10) >DroYak_CAF1 27829 116 - 1 AUUCCAGU-GCCAGCCAGACUGGUCAAACAAG--CCAAAUGCAAAUUUUCAAACCAGACA-UGGGUCUUAACAAGAACAUUUGCAUGAAGUCAAUCAUCUCGACCAUUCUGGCCAGUGAA ....((.(-(...((((((.(((((.......--....((((((((.(((.....((((.-...))))......))).))))))))((......)).....))))).)))))))).)).. ( -33.20) >DroAna_CAF1 28970 115 - 1 GAGAUAGUCGAUAGCCAGUCUGGUCAAACAAGAACCAAAUGCAAAUUUUCUGACCAGACAAUCGGGCUUAACAAGAAAAUUUGCAUGAAGUCAAUCAUCUCGG-----CUGGUCACUGAA .......(((.(..(((((((((((......).)))).(((((((((((((..((........))........)))))))))))))...............))-----))))..).))). ( -29.20) >consensus AUUCCAGU_GCCAGCCAGACUGGUCAAACAAG__CCAAAUGCAAAUUUCCCAACCAGACA_UGGGUCUUAACAAGAACAUUUGCAUGAAGUCAAUCAUCUCGACCAUUCUGGCCAGUGAA .............((((((.(((((.............(((((((((((......((((.....))))......))).))))))))((......)).....))))).))))))....... (-25.28 = -25.70 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:51 2006