| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,884,755 – 10,884,855 |
| Length | 100 |
| Max. P | 0.950993 |

| Location | 10,884,755 – 10,884,855 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -16.35 |
| Energy contribution | -17.35 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
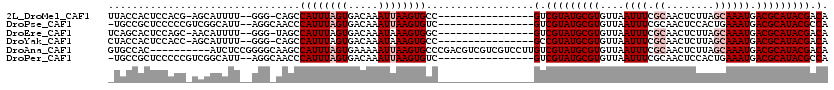
>2L_DroMel_CAF1 10884755 100 + 22407834 UUACCACUCCACG-AGCAUUUU--GGG-CAGCCAUUUAGUGACAAAUUAAGUGCC----------------GUCGUAUGCGUGUUAAUUUCGCAACUCUUAGCAAAUGACGCAUACGACA ...((((((...)-)).....)--)).-..(.((((((((.....)))))))).)----------------(((((((((((....((((.((........)))))).))))))))))). ( -27.70) >DroPse_CAF1 26127 101 + 1 -UGCCGCUCCCCCGUCGGCAUU--AGGCAACCCAUUUAGUGACAAAUUAAGUGUC----------------GUCGUAUGCGUGUUAAUUUCGCAACUCCACUGAAAUGACGCAUACGCCA -((((((......).)))))..--.(((....((((((((.....))))))))..----------------...((((((((....((((((.........)))))).))))))))))). ( -31.50) >DroEre_CAF1 24884 100 + 1 UCAGCACUCCAGC-AACAUUUU--GGG-UAGCCAUUUAGUGACAAAUAAAGUGGC----------------GUCGUAUGCGUGUUAAUUUCGCAACUCUUAGCAAAUGACGCAUACGACA .....((.((((.-......))--)))-).(((((((.((.....)).)))))))----------------(((((((((((....((((.((........)))))).))))))))))). ( -32.20) >DroYak_CAF1 25217 100 + 1 CUACCACUCCACC-AGCAUUUU--GGG-CAGCCAUUUAGUGACAAAUAAAGUGCC----------------GCCGUAUGCGUGUUAAUUUCGCAACUCUUAGCAAAUGACGCAUACGACA .............-.(((((((--(.(-..((......))..)...)))))))).----------------(.(((((((((....((((.((........)))))).))))))))).). ( -23.00) >DroAna_CAF1 26783 110 + 1 GUGCCAC----------AUCUCCGGGGCAAGCCAUUUAGUGAAAAAUUAAGUGCCCGACGUCGUCGUCCUUGUCGUAUGCGUGUUAAUUUCGCAACUCUUAGCAAAUGACGCAUACGACA .((((.(----------......).))))...((((((((.....))))))))...((((....))))..((((((((((((....((((.((........)))))).)))))))))))) ( -34.20) >DroPer_CAF1 26362 101 + 1 -UGCCGCUCCCCCGUCGGCAUU--AGGCAACCCAUUUAGUGACAAAUUAAGUGUC----------------GUCGUAUGCGUGUUAAUUUCGCAACUCCACUGAAAUGACGCAUACGCCA -((((((......).)))))..--.(((....((((((((.....))))))))..----------------...((((((((....((((((.........)))))).))))))))))). ( -31.50) >consensus _UACCACUCCACC_ACCAUUUU__GGG_AAGCCAUUUAGUGACAAAUUAAGUGCC________________GUCGUAUGCGUGUUAAUUUCGCAACUCUUAGCAAAUGACGCAUACGACA ................................((((((((.....))))))))..................(.(((((((((....((((.((........)))))).))))))))).). (-16.35 = -17.35 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:48 2006