| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,884,477 – 10,884,576 |
| Length | 99 |
| Max. P | 0.679826 |

| Location | 10,884,477 – 10,884,576 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -17.79 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10884477 99 + 22407834 UUGGGUUGCGUGCUCGCUUGUUUCGCUU-UUUGUAAUUUUCAAUUUAAGCAGCACAACGUGUUGCUCGC-CAAC----------------UUGCCACAUUGCCACACGGCUACACUA ..((((((((.((.(((((((...((((-.(((.......)))...))))...)))).)))..)).)).-))))----------------))........(((....)))....... ( -23.50) >DroSec_CAF1 24229 100 + 1 UUGGGUUGCGUGCUCGCUUGUUUCGCUU-UUUGUAAUUUUCAAUUUAAGCAGCACAACGUGUUGCUCACCCAAC----------------UUGCCACAUUGCCACAUGGCUACACUU ((((((.(((.((......))..)))..-..................((((((((...)))))))).)))))).----------------..........(((....)))....... ( -24.50) >DroSim_CAF1 24591 100 + 1 UUGGGUUGCGUGCUCGCUUGUUUCGCUU-UUUGUAAUUUUCAAUUUAAGCAGCACAACGUGUUGCUCACCCAAC----------------UUGCCACAUUGCCACAUGGCUACACUU ((((((.(((.((......))..)))..-..................((((((((...)))))))).)))))).----------------..........(((....)))....... ( -24.50) >DroEre_CAF1 24596 112 + 1 UUGGGUUGCGUGCUGGCUUGUUUCGCUU-UUUGUAAUUUUCAAUUUGAGCAACACAACGUGUUGCUCGC-CAAU--G-CUGCUUGCCACAUUGCCACAUUGCCACACGGCUACACUA .(((((.((((..((((.......((..-...))............(((((((((...)))))))))))-))..--)-).))..))).))..........(((....)))....... ( -31.40) >DroYak_CAF1 24923 115 + 1 UUGGGUUGCGUGCCAGCUUGUUUCGCUU-UUUGUAAUUUUCAAUUUAAGCAGCACAACGUGUUGCUCGC-CAACUUGCCUCUUUGCCACAUUGCCACAUGGCCACAUGGCUACACUA ..((((((((.((..((((((...((((-.(((.......)))...))))...)))).))...)).)).-))))))........((((....(((....)))....))))....... ( -27.50) >DroAna_CAF1 26655 92 + 1 CAUCGUUGCGUGCUGGUUUGUUUCGCUUUUUUGUAAUUUUCAAUUUAAGCUACACAACGUGUUGC-------------------------UUGCCACAUUGCCACAUGGCAACUGGU ((.(((((.(((((...(((..(..(......)..)....)))....))).)).)))))))....-------------------------..((((..(((((....))))).)))) ( -21.40) >consensus UUGGGUUGCGUGCUCGCUUGUUUCGCUU_UUUGUAAUUUUCAAUUUAAGCAGCACAACGUGUUGCUCGC_CAAC________________UUGCCACAUUGCCACAUGGCUACACUA ...(((.(((.((......))..))).....................((((((((...))))))))..........................))).....(((....)))....... (-17.79 = -18.07 + 0.28)

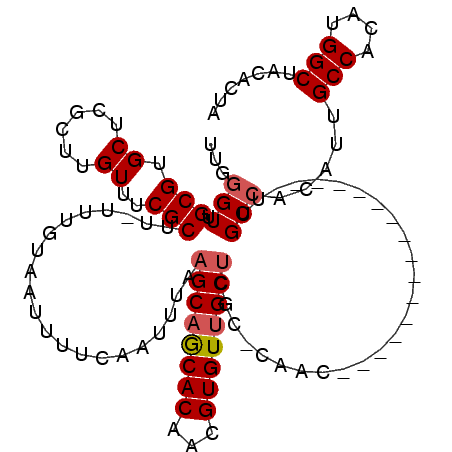

| Location | 10,884,477 – 10,884,576 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.53 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -18.74 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10884477 99 - 22407834 UAGUGUAGCCGUGUGGCAAUGUGGCAA----------------GUUG-GCGAGCAACACGUUGUGCUGCUUAAAUUGAAAAUUACAAA-AAGCGAAACAAGCGAGCACGCAACCCAA ..((((.((((..(....)..))))..----------------(((.-...))).))))((((((.(((((...(((.......))).-..((.......))))))))))))).... ( -26.10) >DroSec_CAF1 24229 100 - 1 AAGUGUAGCCAUGUGGCAAUGUGGCAA----------------GUUGGGUGAGCAACACGUUGUGCUGCUUAAAUUGAAAAUUACAAA-AAGCGAAACAAGCGAGCACGCAACCCAA .......(((((((....)))))))..----------------.(((((((.....)))((((((.(((((...(((.......))).-..((.......))))))))))))))))) ( -27.30) >DroSim_CAF1 24591 100 - 1 AAGUGUAGCCAUGUGGCAAUGUGGCAA----------------GUUGGGUGAGCAACACGUUGUGCUGCUUAAAUUGAAAAUUACAAA-AAGCGAAACAAGCGAGCACGCAACCCAA .......(((((((....)))))))..----------------.(((((((.....)))((((((.(((((...(((.......))).-..((.......))))))))))))))))) ( -27.30) >DroEre_CAF1 24596 112 - 1 UAGUGUAGCCGUGUGGCAAUGUGGCAAUGUGGCAAGCAG-C--AUUG-GCGAGCAACACGUUGUGUUGCUCAAAUUGAAAAUUACAAA-AAGCGAAACAAGCCAGCACGCAACCCAA ..(.((.((.(((((((.....(.((((((.(....).)-)--))))-.)(((((((((...))))))))).................-...........)))).))))).)).).. ( -35.20) >DroYak_CAF1 24923 115 - 1 UAGUGUAGCCAUGUGGCCAUGUGGCAAUGUGGCAAAGAGGCAAGUUG-GCGAGCAACACGUUGUGCUGCUUAAAUUGAAAAUUACAAA-AAGCGAAACAAGCUGGCACGCAACCCAA ..((((.(((((((.(((....))).)))))))..........((..-(((((((.(((...))).)))))...(((.......))).-...........))..))))))....... ( -33.20) >DroAna_CAF1 26655 92 - 1 ACCAGUUGCCAUGUGGCAAUGUGGCAA-------------------------GCAACACGUUGUGUAGCUUAAAUUGAAAAUUACAAAAAAGCGAAACAAACCAGCACGCAACGAUG .....(((((((((....)))))))))-------------------------......((((((((.((((...(((.......)))..))))(........)...))))))))... ( -22.50) >consensus AAGUGUAGCCAUGUGGCAAUGUGGCAA________________GUUG_GCGAGCAACACGUUGUGCUGCUUAAAUUGAAAAUUACAAA_AAGCGAAACAAGCGAGCACGCAACCCAA ..((((.((((..(....)..))))..................(((.....))).))))((((((.((((....(((.......)))....((.......)).)))))))))).... (-18.74 = -19.58 + 0.85)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:47 2006