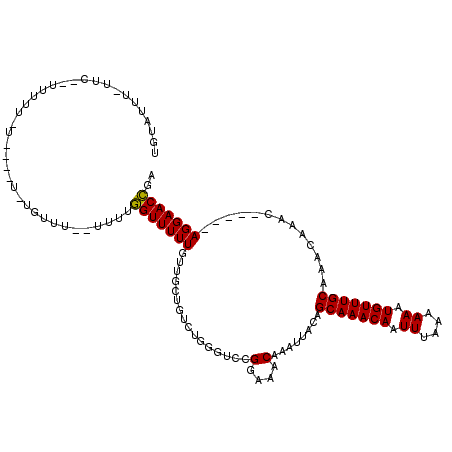
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,869,018 – 10,869,131 |
| Length | 113 |
| Max. P | 0.806196 |

| Location | 10,869,018 – 10,869,131 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -10.33 |
| Energy contribution | -10.57 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10869018 113 + 22407834 UCGGUUCCU-----GUUUGUUUGCAAACAUUUUUAAAAUUGUUUGCUGUAAUUUGUUUCCGGACCCAGACAGCAACAAAAACCAAAA--AAAAAAACCUUAAAAAAAGGGAAUAUAUACA ..((((..(-----(((((....)))))).........(((((.(((((.....(((....)))....)))))))))).))))....--.......((((.....))))........... ( -22.50) >DroSim_CAF1 16174 104 + 1 UCGGUUCCU-----GUUUGUUUGCAAACAUUUUUUAAAUUGUUUGCUGUAAUUUGUUUCCGGACCCAGACAGCAACAAAAACCCAAA--AA----A----AAAAGAACGGAA-AAAUACA ((.((((.(-----(((((....)))))).........(((((.(((((.....(((....)))....)))))))))).........--..----.----....)))).)).-....... ( -21.80) >DroEre_CAF1 16183 99 + 1 UCGGUUCCU-----GUUUGUUUGCAAACAUUUUUUAAAUUGUUUGCUGUAAUUUGUUUCCGGACCCAGACAACAACAAAAACCAAAA--ACACA--------AAAAA------CGAAAAC ..((((..(-----(((((((((((((((..........)))))))........(((....)))..)))))..))))..))))....--.....--------.....------....... ( -16.40) >DroWil_CAF1 20150 111 + 1 UCCGUUCCUGUUUUGUUUGUUUGCAAACAUUUUUUAAAUUGUUUGCCGUAAUUUGUUUCCGGGCCAAAAAACGGGCAAAAACAACAA--CAAC--A----AUAACAAGAGAA-ACAUACA ...((...((((((.((((((.(((((((..........))))))).((...((((((....(((........)))..))))))..)--)...--.----..)))))).)))-))).)). ( -22.80) >DroYak_CAF1 16214 101 + 1 UCGGUUCCU-----GUUUGUUUGCAAACAUUUUUUAAAUUGUUUGCUGUAAUUUGUUUCCGCACCCACACAACAAGAAAAACCAUAAAAAACCA--------AAAAA------AAAUUUU ..((((...-----.((((((.(((((((..........)))))))(((....(((....)))...))).))))))...))))...........--------.....------....... ( -14.20) >consensus UCGGUUCCU_____GUUUGUUUGCAAACAUUUUUUAAAUUGUUUGCUGUAAUUUGUUUCCGGACCCAGACAACAACAAAAACCAAAA__AAACA_A____A_AAAAA__GAA_AAAUACA ..((.(((..............(((((((..........)))))))..............))).))...................................................... (-10.33 = -10.57 + 0.24)

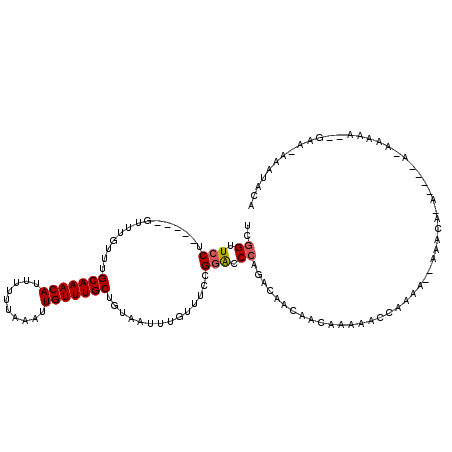
| Location | 10,869,018 – 10,869,131 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.58 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10869018 113 - 22407834 UGUAUAUAUUCCCUUUUUUUAAGGUUUUUUU--UUUUGGUUUUUGUUGCUGUCUGGGUCCGGAAACAAAUUACAGCAAACAAUUUUAAAAAUGUUUGCAAACAAAC-----AGGAACCGA ...........((((.....)))).......--..((((((((((((....((((....))))...........(((((((.((....)).))))))).....)))-----))))))))) ( -25.80) >DroSim_CAF1 16174 104 - 1 UGUAUUU-UUCCGUUCUUUU----U----UU--UUUGGGUUUUUGUUGCUGUCUGGGUCCGGAAACAAAUUACAGCAAACAAUUUAAAAAAUGUUUGCAAACAAAC-----AGGAACCGA .......-.((.(((((...----(----((--((((((((....(((((((..(....)(....).....)))))))..)))))))))))((((((....)))))-----)))))).)) ( -23.50) >DroEre_CAF1 16183 99 - 1 GUUUUCG------UUUUU--------UGUGU--UUUUGGUUUUUGUUGUUGUCUGGGUCCGGAAACAAAUUACAGCAAACAAUUUAAAAAAUGUUUGCAAACAAAC-----AGGAACCGA .......------.....--------.....--..(((((((((((((((.((((....))))...........(((((((.((....)).))))))).))).)))-----))))))))) ( -24.80) >DroWil_CAF1 20150 111 - 1 UGUAUGU-UUCUCUUGUUAU----U--GUUG--UUGUUGUUUUUGCCCGUUUUUUGGCCCGGAAACAAAUUACGGCAAACAAUUUAAAAAAUGUUUGCAAACAAACAAAACAGGAACGGA .......-...(((((((.(----(--((((--(..(((((((((.(((.....)))..)))))))))...))))))).............((((((....)))))).)))))))..... ( -27.10) >DroYak_CAF1 16214 101 - 1 AAAAUUU------UUUUU--------UGGUUUUUUAUGGUUUUUCUUGUUGUGUGGGUGCGGAAACAAAUUACAGCAAACAAUUUAAAAAAUGUUUGCAAACAAAC-----AGGAACCGA .......------.....--------..........((((...(((((((.(((((....(....)...)))))(((((((.((....)).))))))).....)))-----)))))))). ( -20.70) >consensus UGUAUUU_UUC__UUUUU_U____U_UGUUU__UUUUGGUUUUUGUUGCUGUCUGGGUCCGGAAACAAAUUACAGCAAACAAUUUAAAAAAUGUUUGCAAACAAAC_____AGGAACCGA .....................................(((((((................(....)........(((((((.((....)).))))))).............))))))).. (-14.90 = -14.58 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:42 2006