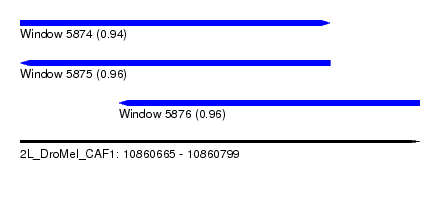
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,860,665 – 10,860,799 |
| Length | 134 |
| Max. P | 0.962864 |

| Location | 10,860,665 – 10,860,769 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
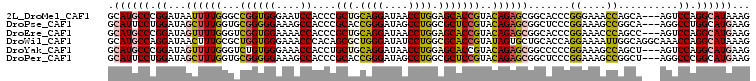
>2L_DroMel_CAF1 10860665 104 + 22407834 CUUUAUGCCUGGACU---UGCUGGUUUCCCGGGUGCCGCUCUGUACGGUGCUCCAGGUUAUCCUGCAGCGGGUGGAUUCCCACCGGCCCAAAAUUAUCCGGGCAUGC ...((((((((((..---(((.((...((.(((..(((.......)))..).)).))....)).)))((.(((((....))))).)).........)))))))))). ( -47.70) >DroPse_CAF1 9196 104 + 1 CUUCAUGCCAGGCCU---UGCCGGCUUUCCGGGAGCCGCUCUGUACGGAGCGCCAGGCUAUCCCGGUGCGGGUGGCUUUCCCCCGCACCAAAGCUAUCCAGGAAUGC ......(((.(((..---.)))))).((((((((((((((((....)))))....)))..))))((((((((.((....))))))))))...........))))... ( -47.80) >DroEre_CAF1 9233 104 + 1 CUUCAUGCCUGGACU---GGCUGGGUUUCCGGGUGCCGCUCUGUACGGUGCUCCAGGUUAUCCUGCAGCGGGUGGUUUUCCACCGACCCAAAACUAUCCGGGCAUGC ...((((((((((..---(..((((((...(((..(((.......)))..)))((((....)))).....(((((....)))))))))))...)..)))))))))). ( -49.10) >DroWil_CAF1 11084 107 + 1 CUUUAUGCCUGGUUUGCCUGCCAAUUUUCCUGGUGCAGCACUAUACGGUGCGCCAGGAUAUCCCAGCGCUGUGGGUUUCCCACCAGCGCAAAGUUAUCCUGGCAUGC ...((((((.((...((..........((((((((((.(.......).)))))))))).......((((((((((...)))).))))))...))...)).)))))). ( -47.80) >DroYak_CAF1 9326 104 + 1 CUUCAUGCCUGGACU---AGCUGGCUUUCCGGGGGCCGCUCUGUACGGUGCUCCAGGUUAUCCUGCAGCAGGUGGUUUCCCACAGACCCAAAACUAUCCGGGCAUGC ...((((((((((.(---((.(((.(((..((..((((((........((((.((((....)))).))))))))))..))...))).)))...))))))))))))). ( -45.00) >DroPer_CAF1 9289 104 + 1 CUUCAUGCCGGGCCU---AGCCGGCUUUCCGGGAGCCGCUCUGUACGGAGCGCCAGGCUAUCCCGGUGCGGGUGGCUUUCCCCCGCACCAAAGCUAUCCAGGAAUGC ...(((.(((((..(---(((((((((.....)))))(((((....)))))....))))).)))((((((((.((....))))))))))...........)).))). ( -50.60) >consensus CUUCAUGCCUGGACU___UGCCGGCUUUCCGGGUGCCGCUCUGUACGGUGCGCCAGGUUAUCCCGCAGCGGGUGGUUUCCCACCGACCCAAAACUAUCCGGGCAUGC ...((((((.(((.......((((....))))(..(((.......)))..))))...............(((((((((............))))))))).)))))). (-23.03 = -23.37 + 0.34)
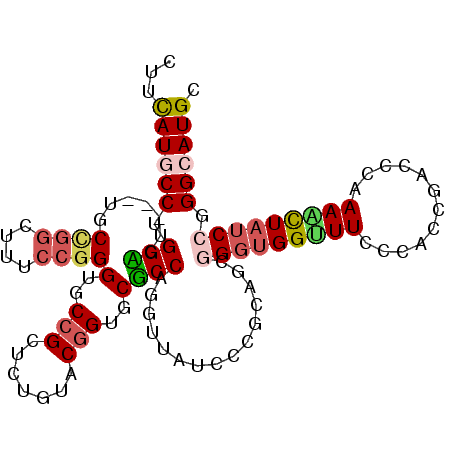
| Location | 10,860,665 – 10,860,769 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -48.05 |
| Consensus MFE | -30.27 |
| Energy contribution | -30.13 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10860665 104 - 22407834 GCAUGCCCGGAUAAUUUUGGGCCGGUGGGAAUCCACCCGCUGCAGGAUAACCUGGAGCACCGUACAGAGCGGCACCCGGGAAACCAGCA---AGUCCAGGCAUAAAG ..(((((.((((......(((..(((((....))))).(((.((((....)))).))).((((.....))))..)))((....))....---.)))).))))).... ( -45.40) >DroPse_CAF1 9196 104 - 1 GCAUUCCUGGAUAGCUUUGGUGCGGGGGAAAGCCACCCGCACCGGGAUAGCCUGGCGCUCCGUACAGAGCGGCUCCCGGAAAGCCGGCA---AGGCCUGGCAUGAAG ...((((.(((..(((((((((((((((....)).))))))))))...)))..(.(((((......))))).)))).)))).((((((.---..))).)))...... ( -48.20) >DroEre_CAF1 9233 104 - 1 GCAUGCCCGGAUAGUUUUGGGUCGGUGGAAAACCACCCGCUGCAGGAUAACCUGGAGCACCGUACAGAGCGGCACCCGGAAACCCAGCC---AGUCCAGGCAUGAAG .((((((.((((.(((((((((.(((((....))))).(((.((((....)))).))))))...))))))(((....(....)...)))---.)))).))))))... ( -47.30) >DroWil_CAF1 11084 107 - 1 GCAUGCCAGGAUAACUUUGCGCUGGUGGGAAACCCACAGCGCUGGGAUAUCCUGGCGCACCGUAUAGUGCUGCACCAGGAAAAUUGGCAGGCAAACCAGGCAUAAAG ((..(((((((((.((..((((((..((....))..))))))..)).)))))))))((((......))))(((.((((.....))))...)))......))...... ( -49.80) >DroYak_CAF1 9326 104 - 1 GCAUGCCCGGAUAGUUUUGGGUCUGUGGGAAACCACCUGCUGCAGGAUAACCUGGAGCACCGUACAGAGCGGCCCCCGGAAAGCCAGCU---AGUCCAGGCAUGAAG .((((((.((((((((..(((((.((((....)))).((((.((((....)))).))))...........)))))..((....))))))---.)))).))))))... ( -46.90) >DroPer_CAF1 9289 104 - 1 GCAUUCCUGGAUAGCUUUGGUGCGGGGGAAAGCCACCCGCACCGGGAUAGCCUGGCGCUCCGUACAGAGCGGCUCCCGGAAAGCCGGCU---AGGCCCGGCAUGAAG .(((.((.((.....(((((((((((((....)).)))))))))))...(((((((((((......))))((((.......)))).)))---)))))))).)))... ( -50.70) >consensus GCAUGCCCGGAUAGCUUUGGGCCGGUGGAAAACCACCCGCUCCAGGAUAACCUGGAGCACCGUACAGAGCGGCACCCGGAAAGCCAGCA___AGUCCAGGCAUGAAG .((((((.((...((((((...((((((....))....(((.((((....)))).)))))))..)))))).......((....))..........)).))))))... (-30.27 = -30.13 + -0.14)
| Location | 10,860,698 – 10,860,799 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -43.63 |
| Consensus MFE | -26.28 |
| Energy contribution | -27.48 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
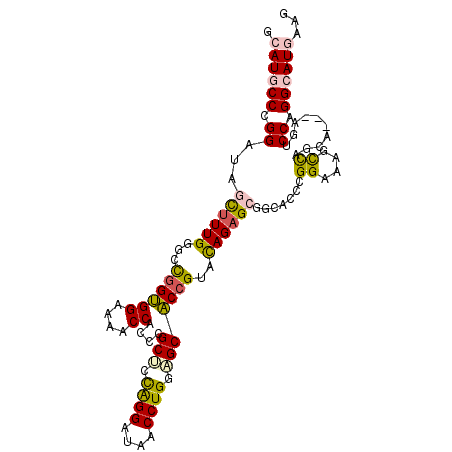
>2L_DroMel_CAF1 10860698 101 - 22407834 GAUUUGGUACGGGGCCAUAUGGCAUCGAGUGCAUGCCCGGAUAAUUUUGGGCCGGUGGGAAUCCACCCGCUGCAGGAUAACCUGGAGCACCGUACAGAGCG ......((((((((((....(((((.......)))))((((....))))))))(((((....))))).(((.((((....)))).))).))))))...... ( -43.00) >DroVir_CAF1 7771 98 - 1 GC---UGUGGCGGUGUAUACGGCAUUGAGUGCAUGCCCGGAUAGGUUUGGCCUGGCGGGAAGCCAGCAGCACUAGGAUAACCAGGUGCGCCGUAGAGCGCA ((---....)).((((.((((((..((...((...((((..((((.....)))).))))..))...))(((((.((....)).)))))))))))..)))). ( -40.60) >DroPse_CAF1 9229 101 - 1 CGUUGGGUACGGGGGCAUAAGGGAGCGAGUGCAUUCCUGGAUAGCUUUGGUGCGGGGGAAAGCCACCCGCACCGGGAUAGCCUGGCGCUCCGUACAGAGCG ((((..((((((((((...(((((((....)).)))))((.((..(((((((((((((....)).))))))))))).)).))..)).))))))))..)))) ( -49.50) >DroEre_CAF1 9266 101 - 1 CAUUUGGUACGGGGCCAUAUGGCAUCGAGUGCAUGCCCGGAUAGUUUUGGGUCGGUGGAAAACCACCCGCUGCAGGAUAACCUGGAGCACCGUACAGAGCG ......((((((.(((....)))...........(((((((....))))))).(((((....))))).(((.((((....)))).))).))))))...... ( -40.70) >DroMoj_CAF1 10925 98 - 1 CU---GGUGGUGGUGCAUACGGCAGAGAGUGCAUGCCCGGAUAGCUUUGGCCUGGCGGAAAGCCCGCUGCGCUGGGAUAACCAGGUGCGCCAUACAGUGCA ((---(...(((((((((...((((.(.((...((((.((..........)).))))....)).).)))).((((.....))))))))))))).))).... ( -38.50) >DroPer_CAF1 9322 101 - 1 CGUUGGGUACGGGGGCAUAAGGGAGCGAGUGCAUUCCUGGAUAGCUUUGGUGCGGGGGAAAGCCACCCGCACCGGGAUAGCCUGGCGCUCCGUACAGAGCG ((((..((((((((((...(((((((....)).)))))((.((..(((((((((((((....)).))))))))))).)).))..)).))))))))..)))) ( -49.50) >consensus CAUU_GGUACGGGGGCAUAAGGCAGCGAGUGCAUGCCCGGAUAGCUUUGGCCCGGCGGAAAGCCACCCGCACCAGGAUAACCUGGAGCACCGUACAGAGCG .........(((((((((.((....)).))))..)))))....((((((...((((((....))....((((((((....))))))))))))..)))))). (-26.28 = -27.48 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:38 2006