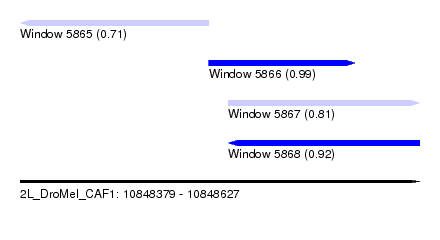
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,848,379 – 10,848,627 |
| Length | 248 |
| Max. P | 0.990411 |

| Location | 10,848,379 – 10,848,496 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.24 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -19.57 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10848379 117 - 22407834 --U-AUUGCAUAUUUAAUUAAAAAUCGUCAGAAACAAGAACGGCAAGUUCAAGGUUGCCUACGGCCAUGAGAACGUCAAGGCCGAUUCGGAUGUGAACAUUGAUCUGAAGGGCCCCUUGA --.-.((((..((((......))))((((........).)))))))..((((((..((((.(((((.(((.....))).))))).(((((((..........))))))))))).)))))) ( -32.50) >DroVir_CAF1 114104 89 - 1 -------------------------------CAACAAGAACGGCAAGUUCAAGGUUGCGUACGGUCAUGAGAAUGUCAAGGCCGAUUCGGAUGUCAAUAUUGACCUGAAGGGUCCCUUGA -------------------------------...((((...((((..((((.(..((....))..).))))..))))..((((..(((((..((((....))))))))).)))).)))). ( -25.20) >DroPse_CAF1 82830 107 - 1 --C-ACU----------UUCUUUCCGAACAGAAACAAGAACGGCAAGUUCAAGGUUGGCUUCGGCCACGAAUACGUGAAAGCCGAUUCGGACGUCAACAUUGAUCUGAAGGGUCCCUUGA --.-.((----------(((..(((((((........((((.....))))..(((((((....)))(((....)))...))))).)))))).((((....))))..)))))......... ( -28.70) >DroYak_CAF1 84608 120 - 1 ACUAACUUGUAGUACUCUUCAUCUCCAUCAGAAACAAGAACGGCAAGUUCAAGGUUGCCUACGGCCAUGAGAACGUCAAGGCCGAUUCGGAUGUGAACAUUGAUCUGAAGGGCCCCUUGA ....((((((.((..((((..(((.....)))...)))))).))))))((((((..((((.(((((.(((.....))).))))).(((((((..........))))))))))).)))))) ( -37.80) >DroMoj_CAF1 117317 89 - 1 -------------------------------CAACAAGAACGGCAAGUUCAAGGUUUCGUACGGUCAUGAGAAUGUCAAGGCCGAUUCGGAUGUCAACAUUGACCUGAAGGGUCCCUUGA -------------------------------......((((.....))))((((..((...(((((.(((.....))).))))).(((((..((((....))))))))).))..)))).. ( -24.90) >DroPer_CAF1 82650 107 - 1 --C-ACU----------UUCUUUCCGAACAGAAACAAGAACGGCAAGUUCAAGGUUGGCUUCGGCCACGAAUACGUGAAAGCCGAUUCGGACGUCAACAUUGAUCUGAAGGGUCCCUUGA --.-.((----------(((..(((((((........((((.....))))..(((((((....)))(((....)))...))))).)))))).((((....))))..)))))......... ( -28.70) >consensus ____ACU__________UUC_U__C_A_CAGAAACAAGAACGGCAAGUUCAAGGUUGCCUACGGCCAUGAGAACGUCAAGGCCGAUUCGGAUGUCAACAUUGAUCUGAAGGGUCCCUUGA ..................................((((...(((...((((.(((((....))))).))))...)))..((((..(((((..((((....))))))))).)))).)))). (-19.57 = -20.18 + 0.61)

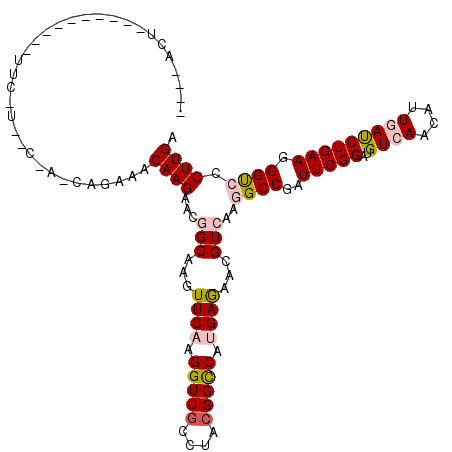
| Location | 10,848,496 – 10,848,587 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.04 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -15.70 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10848496 91 + 22407834 ------------------UUAAUUA-----UUCUGCAGUGAACA-UAUAUGUAAUAACAUGUGAUCCUUACCCAGACUGAGGAGCAAAGUUGCCCUCCAGGGACAGCUUCAGACC ------------------.......-----...(((........-(((((((....))))))).((((((.......)))))))))(((((((((....))).))))))...... ( -22.90) >DroSec_CAF1 81886 91 + 1 ------------------UUAUUUA-----UUCUGCAGGGAACA-CAUAUGUAAUAACAUGUGAUCCUUACCCAGACUGAGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACC ------------------...((((-----.((((.(((((...-(((((((....))))))).)))))...)))).))))....((((((((((....))).)))))))..... ( -29.70) >DroSim_CAF1 88990 87 + 1 ------------------UUA---------UUCUGCAGAGAACA-CAUAUGUAAUAACAUCUGAUCCUUACCCAGACUGAGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACC ------------------...---------.(((((........-((.((((....)))).)).((((((.......))))))))((((((((((....))).)))))))))).. ( -25.20) >DroEre_CAF1 82887 103 + 1 UUGAACUACUGAAUAAUAUGAAUUA-----UU-CGCAGAUAAAAUAAAAUGUAUC------UAUUUCUUACCCAGACUGGGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACC ..........(((((((....))))-----))-)(((((((..........))))------)........(((.....)))..))((((((((((....))).)))))))..... ( -27.70) >DroYak_CAF1 84767 112 + 1 UUUAACUAUUUAAUUAUAUAAAUGACUAUUUU-UGCGGUUAUAAGAAUAUGCAAUAACU--UUGUUCUUACCCAGACUGGGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACC ......((((((......))))))........-...((((.((((((((..........--.))))))))(((.....)))....((((((((((....))).))))))).)))) ( -27.40) >DroAna_CAF1 93289 81 + 1 ------------------UUAAAAA-------CUAUAAACAAUA-GAUAGCUACUAA--------UCUUACCCAGACUGGGGAGCGAAGUUGCCCUCCAGCGACAGCUUCAGACC ------------------.......-------((((.....)))-).......(((.--------(((.....))).)))(((((...(((((......))))).)))))..... ( -15.50) >consensus __________________UUAAUUA_____UUCUGCAGAGAACA_AAUAUGUAAUAACA__UGAUCCUUACCCAGACUGAGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACC ................................................................((((((.......))))))..((((((((((....))).)))))))..... (-15.70 = -15.70 + 0.00)

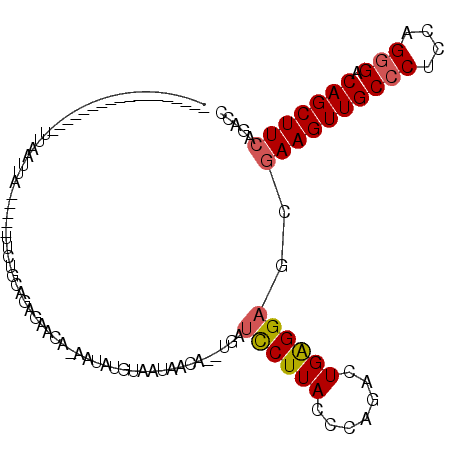
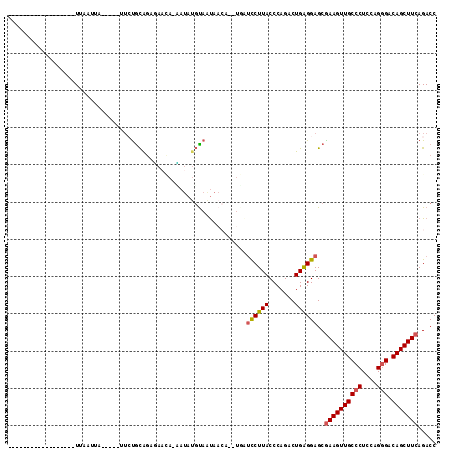
| Location | 10,848,508 – 10,848,627 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -31.94 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10848508 119 + 22407834 CAGUGAACA-UAUAUGUAAUAACAUGUGAUCCUUACCCAGACUGAGGAGCAAAGUUGCCCUCCAGGGACAGCUUCAGACCCUCGAGGAGCUGAUCCUGGACAGCAACCUCGGUGAACAGC ((.(((...-(((((((....))))))).((((((.......)))))).....(((((..(((((((.(((((((.(.....)..))))))).)))))))..))))).))).))...... ( -41.70) >DroSec_CAF1 81898 119 + 1 CAGGGAACA-CAUAUGUAAUAACAUGUGAUCCUUACCCAGACUGAGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACCCUCGAGGAGCUGAUCCUGGACAGCAACCUCGGUGAACAGG .(((((...-(((((((....))))))).))))).......(((...(.(((.(((((..(((((((.(((((((.(.....)..))))))).)))))))..))))).))).)...))). ( -46.90) >DroSim_CAF1 88998 119 + 1 CAGAGAACA-CAUAUGUAAUAACAUCUGAUCCUUACCCAGACUGAGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACCCUCGAGGAGCUGAUCUUGGACAGCAACCUCGGUGAACAGG .......((-(..................((((((.......)))))).(((.(((((..(((((((.(((((((.(.....)..))))))).)))))))..))))).))))))...... ( -39.10) >DroEre_CAF1 82916 114 + 1 CAGAUAAAAUAAAAUGUAUC------UAUUUCUUACCCAGACUGGGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACCCUCGAGGAGCUGAUCCUGGACAGCAACCUCGGUGAACAGC .(((((..........))))------)..(((...(((.....))).(.(((.(((((..(((((((.(((((((.(.....)..))))))).)))))))..))))).))).)))).... ( -39.80) >DroYak_CAF1 84801 118 + 1 CGGUUAUAAGAAUAUGCAAUAACU--UUGUUCUUACCCAGACUGGGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACCCUCGAGGAGCUGAUCCUGGACAGCAACCUCGGUGAACAGC .((((((...........))))))--((((((...(((.....))).(.(((.(((((..(((((((.(((((((.(.....)..))))))).)))))))..))))).))).))))))). ( -44.90) >DroAna_CAF1 93299 111 + 1 UAAACAAUA-GAUAGCUACUAA--------UCUUACCCAGACUGGGGAGCGAAGUUGCCCUCCAGCGACAGCUUCAGACCCUCCAACAGCUGAUCCUGAACAGCAACCUCGGUGAAGAGG .........-....(((.....--------.........(.((((((.(((....))).)))))))..(((..((((............))))..)))...)))..((((......)))) ( -26.40) >consensus CAGAGAACA_AAUAUGUAAUAACA__UGAUCCUUACCCAGACUGAGGAGCGAAGUUGCCCUCCAGGGACAGCUUCAGACCCUCGAGGAGCUGAUCCUGGACAGCAACCUCGGUGAACAGC .............................((((((.......)))))).(((.(((((..(((((((.(((((((..........))))))).)))))))..))))).)))......... (-31.94 = -32.30 + 0.36)

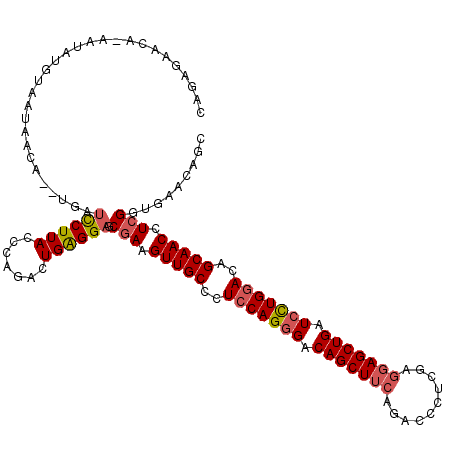
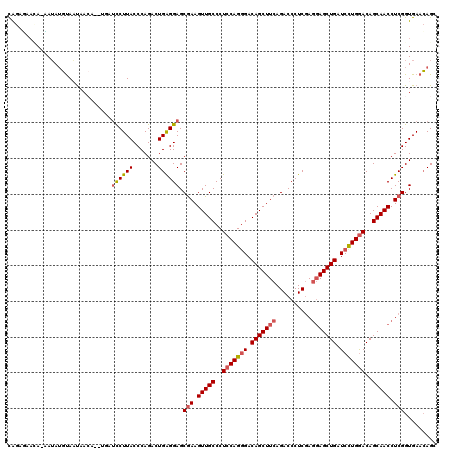
| Location | 10,848,508 – 10,848,627 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -43.67 |
| Consensus MFE | -36.99 |
| Energy contribution | -37.13 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10848508 119 - 22407834 GCUGUUCACCGAGGUUGCUGUCCAGGAUCAGCUCCUCGAGGGUCUGAAGCUGUCCCUGGAGGGCAACUUUGCUCCUCAGUCUGGGUAAGGAUCACAUGUUAUUACAUAUA-UGUUCACUG ((((.....((((((((((.((((((..(((((..(((......))))))))..)))))).)))))))))).....)))).......((((..((((((........)))-))))).)). ( -42.50) >DroSec_CAF1 81898 119 - 1 CCUGUUCACCGAGGUUGCUGUCCAGGAUCAGCUCCUCGAGGGUCUGAAGCUGUCCCUGGAGGGCAACUUCGCUCCUCAGUCUGGGUAAGGAUCACAUGUUAUUACAUAUG-UGUUCCCUG .(((.....((((((((((.((((((..(((((..(((......))))))))..)))))).)))))))))).....))).........(((.(((((((......)))))-)).)))... ( -50.00) >DroSim_CAF1 88998 119 - 1 CCUGUUCACCGAGGUUGCUGUCCAAGAUCAGCUCCUCGAGGGUCUGAAGCUGUCCCUGGAGGGCAACUUCGCUCCUCAGUCUGGGUAAGGAUCAGAUGUUAUUACAUAUG-UGUUCUCUG .......(((((((..((((........)))).))))(((((..(((((.(((((.....))))).))))).)))))......))).(((((((.((((....)))).))-.)))))... ( -38.50) >DroEre_CAF1 82916 114 - 1 GCUGUUCACCGAGGUUGCUGUCCAGGAUCAGCUCCUCGAGGGUCUGAAGCUGUCCCUGGAGGGCAACUUCGCUCCCCAGUCUGGGUAAGAAAUA------GAUACAUUUUAUUUUAUCUG ((((.....((((((((((.((((((..(((((..(((......))))))))..)))))).)))))))))).....))))..((((((..((((------((.....)))))))))))). ( -44.70) >DroYak_CAF1 84801 118 - 1 GCUGUUCACCGAGGUUGCUGUCCAGGAUCAGCUCCUCGAGGGUCUGAAGCUGUCCCUGGAGGGCAACUUCGCUCCCCAGUCUGGGUAAGAACAA--AGUUAUUGCAUAUUCUUAUAACCG ..(((((..((((((((((.((((((..(((((..(((......))))))))..)))))).))))))))))...(((.....)))...))))).--.(((((...........))))).. ( -46.80) >DroAna_CAF1 93299 111 - 1 CCUCUUCACCGAGGUUGCUGUUCAGGAUCAGCUGUUGGAGGGUCUGAAGCUGUCGCUGGAGGGCAACUUCGCUCCCCAGUCUGGGUAAGA--------UUAGUAGCUAUC-UAUUGUUUA ..((((...((((((((((.(((((((.(((((.(..(.....)..)))))))).))))).))))))))))...(((.....))).))))--------.((((((....)-))))).... ( -39.50) >consensus CCUGUUCACCGAGGUUGCUGUCCAGGAUCAGCUCCUCGAGGGUCUGAAGCUGUCCCUGGAGGGCAACUUCGCUCCCCAGUCUGGGUAAGAAUCA__UGUUAUUACAUAUG_UGUUCUCUG ...((((..((((((((((.((((((..(((((..(((......))))))))..)))))).))))))))))...(((.....)))...))))............................ (-36.99 = -37.13 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:31 2006