
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,848,200 – 10,848,339 |
| Length | 139 |
| Max. P | 0.969605 |

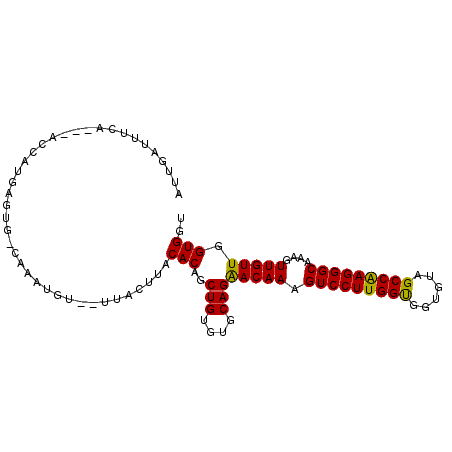
| Location | 10,848,200 – 10,848,299 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -23.86 |
| Energy contribution | -23.33 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10848200 99 + 22407834 AUUGAUUUCAUUUGCCAUGCGUGGCAAUUGCA-AUACUUACACAGCUGUGUGCAGAACAAAGUCCUUGGUGGUGUAGCCAAGGGCAAAGUUGUUGGUGGU ........((.((((((....)))))).))..-.((((...((((((.(((.....)))..(((((((((......)))))))))..)))))).)))).. ( -32.20) >DroGri_CAF1 99541 89 + 1 AUAGAGUUU----GUAAUAC-UG-----UGU-AUUACUUACACGGCUGUGUGCAGGACAAAGUCCUUGGCGGAGUAGCCGAGGGCAAAGUUGUUGGUGGU .......((----(((((((-((-----(((-(.....))))))).))).)))))(((((.(((((((((......)))))))))....)))))...... ( -31.20) >DroEre_CAF1 82552 96 + 1 AUUGAUUUAA---ACCAAGUGUGCCAGAUGUA-AUACUUACACAGCUGUGUGCAGAACAAAGUCCUUGGUGGUGUAGCCAAGGGCAAAGUUGUUGGUGGU ..........---((((..((..((((.((((-.....))))...))).)..)).(((((.(((((((((......)))))))))....)))))..)))) ( -29.90) >DroWil_CAF1 130024 94 + 1 GGAAAUUUCA---AGCAGGACUAACAAAA---UUUACUUACACAGCUGUGUGCAGGACAAAGUCCUUGGUGGUAUAGCCAAGGGCAAAGUUGUUGGUGGU ..........---......((((((((..---....(((((((....))))).))......(((((((((......)))))))))....))))))))... ( -28.50) >DroYak_CAF1 84435 93 + 1 AUUGAUUUUU---GCCAAGUGUGCCAGGUGU----ACUUACACAGCUGUGUGCAGAACAAAGUCCUUGGUGGUGUAGCCAAGGGCAAAGUUGUUGGUGGU ........(.---.((...((..((((.(((----......))).))).)..)).(((((.(((((((((......)))))))))....)))))))..). ( -33.40) >DroAna_CAF1 93009 90 + 1 AUUAAAAGGA---ACAAUGAG-----AGUG-G-UUACUUACACAGCUGUGUGCAGGACAAAGUCCUUGGUGGUGUAGCCAAGGGCAAAGUUGUUGGUGGU .....(((.(---((.((...-----.)).-)-)).))).(((..(((....)))(((((.(((((((((......)))))))))....))))).))).. ( -26.80) >consensus AUUGAUUUCA___ACCAUGAGUG_CAAAUGU__UUACUUACACAGCUGUGUGCAGAACAAAGUCCUUGGUGGUGUAGCCAAGGGCAAAGUUGUUGGUGGU ........................................(((..(((....)))(((((.(((((((((......)))))))))....))))).))).. (-23.86 = -23.33 + -0.53)

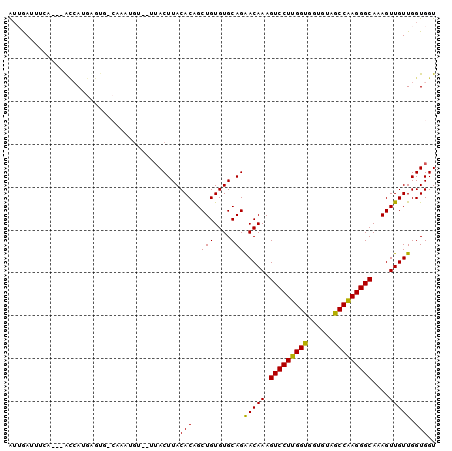
| Location | 10,848,200 – 10,848,299 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -12.70 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10848200 99 - 22407834 ACCACCAACAACUUUGCCCUUGGCUACACCACCAAGGACUUUGUUCUGCACACAGCUGUGUAAGUAU-UGCAAUUGCCACGCAUGGCAAAUGAAAUCAAU .............((((((((((........)))))).........((((((....)))))).....-.))))((((((....))))))........... ( -23.30) >DroGri_CAF1 99541 89 - 1 ACCACCAACAACUUUGCCCUCGGCUACUCCGCCAAGGACUUUGUCCUGCACACAGCCGUGUAAGUAAU-ACA-----CA-GUAUUAC----AAACUCUAU .............((((...(((((.....((..(((((...)))))))....))))).))))(((((-((.-----..-)))))))----......... ( -20.10) >DroEre_CAF1 82552 96 - 1 ACCACCAACAACUUUGCCCUUGGCUACACCACCAAGGACUUUGUUCUGCACACAGCUGUGUAAGUAU-UACAUCUGGCACACUUGGU---UUAAAUCAAU ...(((((......(((((((((........))))).....(((..(((((((....))))..))).-.)))...))))...)))))---.......... ( -19.50) >DroWil_CAF1 130024 94 - 1 ACCACCAACAACUUUGCCCUUGGCUAUACCACCAAGGACUUUGUCCUGCACACAGCUGUGUAAGUAAA---UUUUGUUAGUCCUGCU---UGAAAUUUCC ...((.(((((.(((((((((((........)))))).........((((((....)))))).)))))---..))))).))......---.......... ( -18.60) >DroYak_CAF1 84435 93 - 1 ACCACCAACAACUUUGCCCUUGGCUACACCACCAAGGACUUUGUUCUGCACACAGCUGUGUAAGU----ACACCUGGCACACUUGGC---AAAAAUCAAU ............(((((((((((........))))))...............(((.(((((.((.----....)).))))).)))))---)))....... ( -20.00) >DroAna_CAF1 93009 90 - 1 ACCACCAACAACUUUGCCCUUGGCUACACCACCAAGGACUUUGUCCUGCACACAGCUGUGUAAGUAA-C-CACU-----CUCAUUGU---UCCUUUUAAU .........(((..((....(((.(((.......(((((...)))))(((((....)))))..))).-)-))..-----..))..))---)......... ( -15.80) >consensus ACCACCAACAACUUUGCCCUUGGCUACACCACCAAGGACUUUGUCCUGCACACAGCUGUGUAAGUAA__ACACUUG_CACACAUGGC___UAAAAUCAAU .................((((((........)))))).........(((((......)))))...................................... (-12.70 = -12.87 + 0.17)


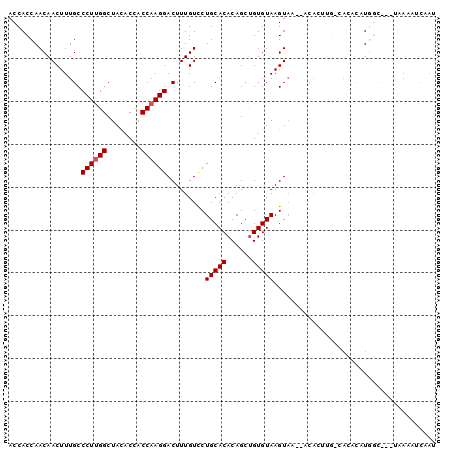
| Location | 10,848,227 – 10,848,339 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.96 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -30.96 |
| Energy contribution | -30.27 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10848227 112 + 22407834 -UUGCA-----AUACUUACACAGCUGUGUGCAGAACAAAGUCCUUGGUGGUGUAGCCAAGGGCAAAGUUGUUGGUGGUCAGCUUGGACUGUUGUGUGUCAAAUGCGGUCUGGUAGCCG -..(((-----.....((((((((...((.(........(((((((((......))))))))).((((((........))))))).)).)))))))).....)))(((......))). ( -38.40) >DroVir_CAF1 113949 115 + 1 --CUG-AUUGUUUACUUACACGGCUGUGUGGAGGACAAAGUCCUUGGUGGAGUAGCCGAGGGCAAAGUUGUUGGUGGUCAACUUGGACUGUUGUGUAUCGAAUGCGGUCUGAUAACCG --..(-(((((.....((((((((.((.(..........(((((((((......))))))))).((((((........))))))).)).))))))))......))))))......... ( -36.30) >DroGri_CAF1 99559 111 + 1 --UGU-----AUUACUUACACGGCUGUGUGCAGGACAAAGUCCUUGGCGGAGUAGCCGAGGGCAAAGUUGUUGGUGGUCAACUUGGACUGUUUUGUAUCGAAUGCGGUCUGAUAGCCG --(((-----(.....))))(((((((((((((((((..(((((((((......))))))))).((((((........))))))....))))))))))((....)).....))))))) ( -42.10) >DroWil_CAF1 130048 110 + 1 -AA-------UUUACUUACACAGCUGUGUGCAGGACAAAGUCCUUGGUGGUAUAGCCAAGGGCAAAGUUGUUGGUGGUCAACUUGGCUUUCUGUGUAUCGAAUGCCGUCUGGUAGCCA -..-------.((((((((((....)))))..((((...(((((((((......))))))))).((((((........))))))(((.(((........))).))))))))))))... ( -36.20) >DroMoj_CAF1 117162 115 + 1 --ACG-AAUUUGUACUUACACGGCUGUGUGCAGGACAAAGUCCUUGGUGGAGUAGCCGAGGGCAAAGUUGUUGGUGGUCAACUUGGAUUGUUGUGUAUCGAAGGCAGUUUGAUAGCCA --...-...............(((((((((((.(((((.(((((((((......))))))))).((((((........))))))...))))).)))))((((.....)))))))))). ( -38.10) >DroPer_CAF1 82492 118 + 1 GGUGCUACUGUUUACUUACACAGAUGUGUGCAAGACAAAGUCCUUGGUGGUGUAGCCAAGGGCAAAGUUGUUGGUGGUCAACUUGGUCUGUUGCGUAUCAAAUGCGGCCUGGUAGCCG .(.((((((.......(((((....)))))..((((...(((((((((......))))))))).((((((........)))))).))))(((((((.....)))))))..))))))). ( -42.30) >consensus __UGC_____UUUACUUACACAGCUGUGUGCAGGACAAAGUCCUUGGUGGAGUAGCCAAGGGCAAAGUUGUUGGUGGUCAACUUGGACUGUUGUGUAUCGAAUGCGGUCUGAUAGCCG ..............(((((((....))))).))(((((.(((((((((......)))))))))....)))))...(((....(..((((((............))))))..)..))). (-30.96 = -30.27 + -0.69)
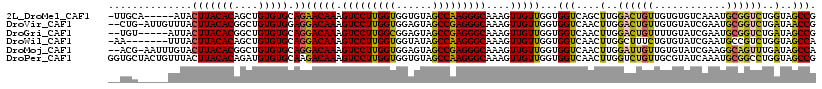

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:27 2006