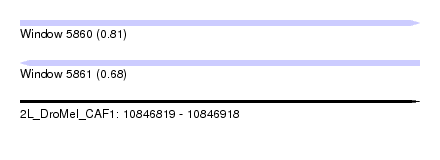
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,846,819 – 10,846,918 |
| Length | 99 |
| Max. P | 0.812788 |

| Location | 10,846,819 – 10,846,918 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -38.41 |
| Consensus MFE | -35.80 |
| Energy contribution | -35.42 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10846819 99 + 22407834 UGCCCGGAUGGUAGCCGCGCUUGAAGAGAUCGCGGGCCAGCUUGCCCAGAUCUGGGUAUGUCGGUGUUUUGGCGGCCAUUACGGGCUGUAUGAAUAGGU .(((((((((((.((((((.((.....)).)))..(((.((.((((((....)))))).)).))).....))).)))))).)))))............. ( -42.00) >DroSec_CAF1 80237 99 + 1 UGCCCGGAUGGUAGCCGCGCUUGAAGAGAUCGCGGGCCAGCUUGCCCAGAUCCGGGUAUGUCGGCGUUUUGGCGGCCAUUGCAGGCGGUAUGAAUAGGU .(((.(((((((.((((((.((.....)).)))..(((.((.(((((......))))).)).))).....))).)))))).).)))............. ( -36.80) >DroSim_CAF1 87354 99 + 1 UGCCCGGAUGGUAGCCGCGCUUGAAGAGAUCGCGGGCCAGCUUGCCCAGAUCCGGGUAUGUCGGCGUUUUGGCGGCCAUUGCGGGCGGUAUGAAUAGGU .(((((((((((.((((((.((.....)).)))..(((.((.(((((......))))).)).))).....))).)))))).)))))............. ( -42.90) >DroEre_CAF1 81215 99 + 1 UGCCCGGAUGGUAGCCGCGCUUGAAGAGAUCGCGGGCCAGCUUGCCCAGGUCAGGGUAUGUCGGCGUGGUGGUGGCCAUUAUGGGCGGAAUGAAUAGGU .(((((((((((.(((((....((.....)).((.(((.((.(((((......))))).)).))).))))))).)))))).)))))............. ( -41.10) >DroYak_CAF1 83093 99 + 1 UGCCCGGAUGGUAGCCGCGCUUGAAGAGAUCGCGGGCCAGCUUGCCCAGGUCGGGGUAUGUAGGCGUGGUUUUGGCCAUUAUGGGCGGUAUGAAGAGGU .((((((((((((((((((((((....((((..((((......)))).))))........)))))))))))...)))))).)))))............. ( -42.60) >DroMoj_CAF1 115867 93 + 1 GGCCAGGAUGAUAACCACGCUUGAACAAAUCGCGGGCCAGUUUGCCCAAAUCAGCAUAAACAGGCGGC------GCCAUUUUUGGCCAAAUAAAUAAUU ((((((((.........((((((..(.....).((((......)))).............))))))..------.....))))))))............ ( -25.09) >consensus UGCCCGGAUGGUAGCCGCGCUUGAAGAGAUCGCGGGCCAGCUUGCCCAGAUCAGGGUAUGUCGGCGUUUUGGCGGCCAUUACGGGCGGUAUGAAUAGGU ((((((((((((..(((((.((.....)).)))))(((.((.(((((......))))).)).))).........)))))).))))))............ (-35.80 = -35.42 + -0.38)

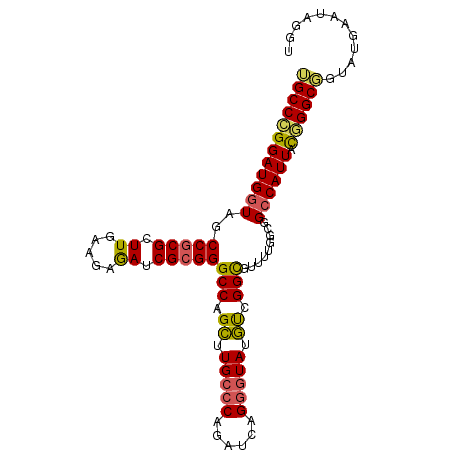
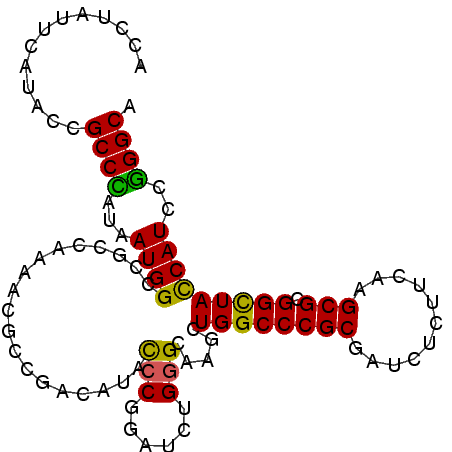
| Location | 10,846,819 – 10,846,918 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -23.95 |
| Energy contribution | -23.32 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10846819 99 - 22407834 ACCUAUUCAUACAGCCCGUAAUGGCCGCCAAAACACCGACAUACCCAGAUCUGGGCAAGCUGGCCCGCGAUCUCUUCAAGCGCGGCUACCAUCCGGGCA .............(((((...((((((((........(.......)((((((((((......))))).)))))......).))))))).....))))). ( -33.90) >DroSec_CAF1 80237 99 - 1 ACCUAUUCAUACCGCCUGCAAUGGCCGCCAAAACGCCGACAUACCCGGAUCUGGGCAAGCUGGCCCGCGAUCUCUUCAAGCGCGGCUACCAUCCGGGCA .............(((((...((((((((.......((.......))(((((((((......))))).)))).......).))))))).....))))). ( -33.00) >DroSim_CAF1 87354 99 - 1 ACCUAUUCAUACCGCCCGCAAUGGCCGCCAAAACGCCGACAUACCCGGAUCUGGGCAAGCUGGCCCGCGAUCUCUUCAAGCGCGGCUACCAUCCGGGCA .............(((((...((((((((.......((.......))(((((((((......))))).)))).......).))))))).....))))). ( -35.20) >DroEre_CAF1 81215 99 - 1 ACCUAUUCAUUCCGCCCAUAAUGGCCACCACCACGCCGACAUACCCUGACCUGGGCAAGCUGGCCCGCGAUCUCUUCAAGCGCGGCUACCAUCCGGGCA .............((((...((((..........((((.(...(((......)))...).))))(((((...........)))))...))))..)))). ( -28.00) >DroYak_CAF1 83093 99 - 1 ACCUCUUCAUACCGCCCAUAAUGGCCAAAACCACGCCUACAUACCCCGACCUGGGCAAGCUGGCCCGCGAUCUCUUCAAGCGCGGCUACCAUCCGGGCA .............((((...((((..........(((((............)))))....((((((((...........))).)))))))))..)))). ( -27.90) >DroMoj_CAF1 115867 93 - 1 AAUUAUUUAUUUGGCCAAAAAUGGC------GCCGCCUGUUUAUGCUGAUUUGGGCAAACUGGCCCGCGAUUUGUUCAAGCGUGGUUAUCAUCCUGGCC ............(((((...((((.------(((((..((((..((.(((((((((......))))).)))).))..)))))))))..))))..))))) ( -27.30) >consensus ACCUAUUCAUACCGCCCAUAAUGGCCGCCAAAACGCCGACAUACCCGGAUCUGGGCAAGCUGGCCCGCGAUCUCUUCAAGCGCGGCUACCAUCCGGGCA .............((((...((((...................(((......))).....((((((((...........))).)))))))))..)))). (-23.95 = -23.32 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:24 2006