| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
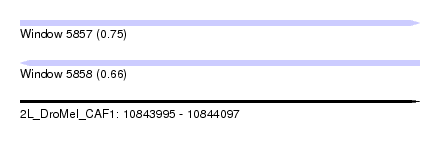
| Location | 10,843,995 – 10,844,097 |
| Length | 102 |
| Max. P | 0.746114 |

| Location | 10,843,995 – 10,844,097 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -25.41 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10843995 102 + 22407834 GCCAUUGAUGAGGCAAUCCUUCGCCAAGGCGCCCACAUGGAAGUAGGUGGGCAUUUCGCCCUCUGACGUUGGCGAGGGUAUUAUCUCCACUUCGACUUCCUU ....((((.(((..(((((((((((((.(((((.((......)).)))((((.....))))...).).)))))))))).)))..)))....))))....... ( -36.60) >DroSec_CAF1 77423 102 + 1 GCCAUUGAUGAGACAAUCCUUCGCCAGGGCGCCCACAUGGAAGUAGGUGGGCAUUUCGCCCUCUGACGUUGGCGAGGGAAUAAUCUCCACUUCGACUUCCUU ....((((.((((...((((((((((((((((((((..........)))))).....))))........))))))))))....))))....))))....... ( -36.90) >DroSim_CAF1 84525 102 + 1 GCCAUUGAUGAGACAAUCCUUCGCCAAGGCGCCCACAUGGAAGUAGGUGGGCAUUUCGCCCUCUGCCGUUGGCGAGGGUAUAAUCUCCACUUCGACUUCCUU (((.(((.((((.......)))).))))))........((((((.((((((.(((..((((((.(((...)))))))))..))).))))))...)))))).. ( -39.50) >DroEre_CAF1 78305 102 + 1 ACCAUUGAUAAGGCAUUCCUUCGCCAAGGAGCCCACAUGGAAGUAGGAGGGCAUUUCGCCCUCUGCCGGUGGCGCGGGUAUUAUCUCUAUAUCGAUUUCCUU ...........(((........)))(((((((((((.(((.....(((((((.....))))))).))))))).)).(((((.......)))))....))))) ( -36.20) >DroYak_CAF1 80175 102 + 1 ACCAUUGAUAAGGCAAUCCUUUGCCAAGGCGCCCACAUGAAAGUAGGUGGGCAUUUCGCCCUCCGCCGGUGGCAUGGGUAUUAUCUCUACCUCGACAUCCUU ......(((((....((((..(((((.((((((.((......)).)))((((.....))))...)))..))))).)))).)))))................. ( -31.90) >DroPer_CAF1 79667 102 + 1 GCCCCUGAUGAGACACUCCUUGGCCGCACCACCCACGUGGAAAUAUGUGGGCAUCUCGCCCUCGGGCACCGGCACCGGCAGUAUCUCUACGUCAUCCUUCAC .....((((((((.(((((...((((.....((((((((....))))))))......(((....)))..))))...)).))).))))...))))........ ( -35.00) >consensus GCCAUUGAUGAGACAAUCCUUCGCCAAGGCGCCCACAUGGAAGUAGGUGGGCAUUUCGCCCUCUGCCGUUGGCGAGGGUAUUAUCUCCACUUCGACUUCCUU ....((((.((((.((((((.(((((......((....)).....((.((((.....)))).)).....))))).))).))).))))....))))....... (-25.41 = -25.42 + 0.00)

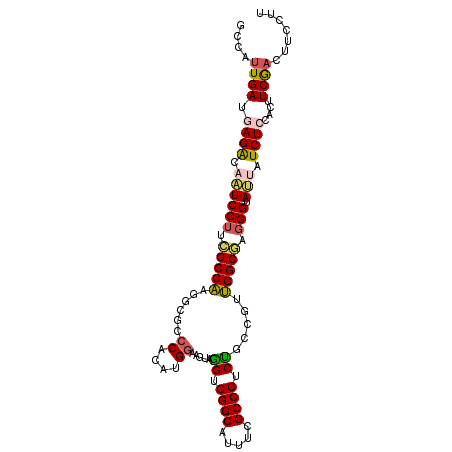
| Location | 10,843,995 – 10,844,097 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.15 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10843995 102 - 22407834 AAGGAAGUCGAAGUGGAGAUAAUACCCUCGCCAACGUCAGAGGGCGAAAUGCCCACCUACUUCCAUGUGGGCGCCUUGGCGAAGGAUUGCCUCAUCAAUGGC ......((((.....(((.((((.((.(((((((.((....((((.....)))).(((((......))))).)).))))))).)))))).))).....)))) ( -35.40) >DroSec_CAF1 77423 102 - 1 AAGGAAGUCGAAGUGGAGAUUAUUCCCUCGCCAACGUCAGAGGGCGAAAUGCCCACCUACUUCCAUGUGGGCGCCCUGGCGAAGGAUUGUCUCAUCAAUGGC ......((((.....(((((...(((.(((((..(....)(((((.....((((((..........)))))))))))))))).)))..))))).....)))) ( -38.20) >DroSim_CAF1 84525 102 - 1 AAGGAAGUCGAAGUGGAGAUUAUACCCUCGCCAACGGCAGAGGGCGAAAUGCCCACCUACUUCCAUGUGGGCGCCUUGGCGAAGGAUUGUCUCAUCAAUGGC ......((((.....(((((.((.((.(((((((.(((...((((.....)))).(((((......))))).)))))))))).)))).))))).....)))) ( -42.40) >DroEre_CAF1 78305 102 - 1 AAGGAAAUCGAUAUAGAGAUAAUACCCGCGCCACCGGCAGAGGGCGAAAUGCCCUCCUACUUCCAUGUGGGCUCCUUGGCGAAGGAAUGCCUUAUCAAUGGU ((((..(((........)))....((..(((((..(((.((((((.....))))))((((......)))))))...)))))..))....))))......... ( -32.60) >DroYak_CAF1 80175 102 - 1 AAGGAUGUCGAGGUAGAGAUAAUACCCAUGCCACCGGCGGAGGGCGAAAUGCCCACCUACUUUCAUGUGGGCGCCUUGGCAAAGGAUUGCCUUAUCAAUGGU ((((..((((.((((.......))))).(((((..((((..((((.....)))).(((((......))))))))).)))))...)))..))))......... ( -37.20) >DroPer_CAF1 79667 102 - 1 GUGAAGGAUGACGUAGAGAUACUGCCGGUGCCGGUGCCCGAGGGCGAGAUGCCCACAUAUUUCCACGUGGGUGGUGCGGCCAAGGAGUGUCUCAUCAGGGGC ...............((((((((.((...((((.((((....))))....((((((..........))))))....))))...))))))))))......... ( -37.20) >consensus AAGGAAGUCGAAGUAGAGAUAAUACCCUCGCCAACGGCAGAGGGCGAAAUGCCCACCUACUUCCAUGUGGGCGCCUUGGCGAAGGAUUGCCUCAUCAAUGGC ...............(((.((((.((..(((((..(((...((((.....)))).(((((......))))).))).)))))..)))))).)))......... (-26.20 = -26.15 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:21 2006