| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,818,176 – 10,818,277 |
| Length | 101 |
| Max. P | 0.570261 |

| Location | 10,818,176 – 10,818,277 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.59 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -14.38 |
| Energy contribution | -14.45 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
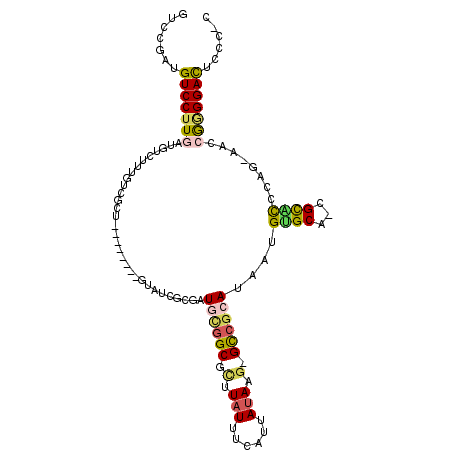
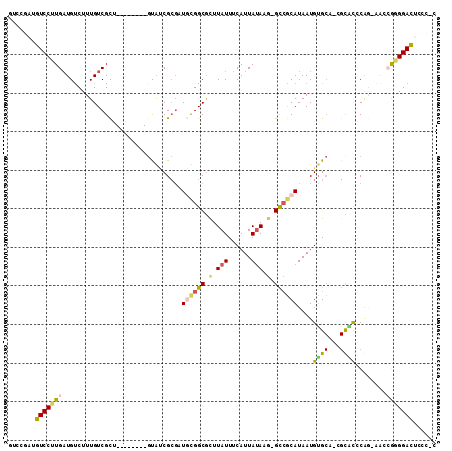
>2L_DroMel_CAF1 10818176 101 - 22407834 GUCCGAUGUCCUUGAUGUCUUUGUCGCU--------GUAUCGCGAUGUGGCGCUUAUUUCAUUAUAAG-GCCGCAUAAUGUGCAGAGCACCCGGCAACCGGGGACUCCC-C ((((((((((...))))))..(((..((--------((((....(((((((.(((((......)))))-)))))))...)))))).))).((((...))))))))....-. ( -36.40) >DroVir_CAF1 67461 94 - 1 GCCGCAAGUCCUUGCUGCCUUUGUCGC---------GUGCCGCGUUCCGGCAUUUAUUUCAUUACAU--GCAACAUAAUGGGCG-CGUUUCCAU-----ACGGAUACUUUC ((.((((....)))).)).......((---------(((((.((((...((((.((......)).))--)).....))))))))-))).(((..-----..)))....... ( -25.30) >DroEre_CAF1 52310 101 - 1 GUCCGAUGUCCUUGAUGUCUUUGUCGCU--------GUAUCGCGAUGUGGCGCUUAUUUCAUUAUAAG-GCCGCAUAAUGUGCAGCGCACCCAGCAACCGGGGACUCCC-C ....((.(((((((.((.((..((((((--------((((....(((((((.(((((......)))))-)))))))...)))))))).))..))))..)))))))))..-. ( -39.20) >DroWil_CAF1 78749 101 - 1 ---UAAUGUCCUUGAUGUCUUUGUCGUUUGGCUUUAGCGGCGCGUUCCGGCGUUUAUUUCAUUAUAUGGGCCGCAUAAUGUGCG-CGUUUCCAU-----AAGGACUUCU-C ---....(((((((((......)))...(((....(((((((((((.((((.(.(((......))).).))))...))))))).-)))).))).-----))))))....-. ( -29.90) >DroYak_CAF1 53742 101 - 1 GUCCGAUGUCCUUGAUGUCUUUGUCGCU--------GUAUCGCGAUGUGGCGCUUAUUUCAUUAUAAG-GCCGCAUAAUGUGCAGAGCACCCGGGAACCGGGGACUCCC-C ((((((((((...))))))..(((..((--------((((....(((((((.(((((......)))))-)))))))...)))))).))).((((...))))))))....-. ( -36.40) >DroAna_CAF1 53491 91 - 1 ---UAAUGUCCUUGAUGUCUUUGUCGCU--------UUGUCG--UUGCGGCGCUUAUUUCAUUAUAUG-GUCGCAUAAUGUG-----CGCCCGGAAAACGGGGACCCUU-C ---....(((((((...(((..(((((.--------......--..)))))................(-(.(((((...)))-----)).)))))...)))))))....-. ( -25.70) >consensus GUCCGAUGUCCUUGAUGUCUUUGUCGCU________GUAUCGCGAUGCGGCGCUUAUUUCAUUAUAAG_GCCGCAUAAUGUGCA_CGCACCCAG_AACCGGGGACUCCC_C .......(((((((...............................((((((.(.(((......))).).))))))....((((...))))........)))))))...... (-14.38 = -14.45 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:13 2006