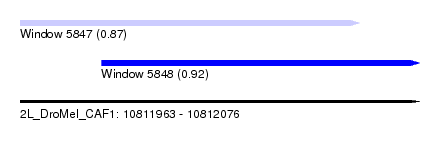
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,811,963 – 10,812,076 |
| Length | 113 |
| Max. P | 0.919809 |

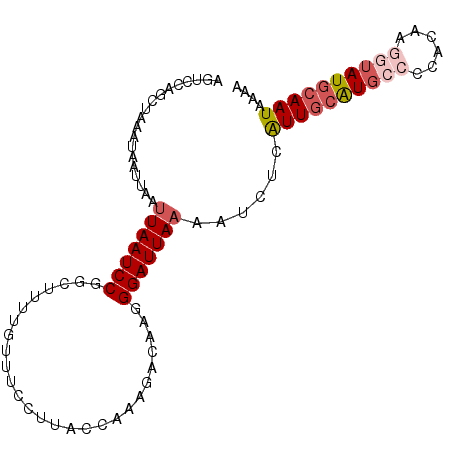
| Location | 10,811,963 – 10,812,059 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -19.25 |
| Consensus MFE | -12.59 |
| Energy contribution | -13.65 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10811963 96 + 22407834 AGUCCAGCUAAAUAAUUAAUUAAUCCGGCUUUCGUUUCGUUACCAAAGACAAGGGAUUAAAAUCUCAUUGCAUGGCCCACAAGGUAUGCAAUAAAG ...................(((((((.(((((.((......)).)))).)...)))))))......((((((((.((.....)))))))))).... ( -18.50) >DroSec_CAF1 45607 94 + 1 --UCCAGCUAAAUAAUUAAUUAAUCCGGCUUUUGUUUCCUUACCACAGACAAGGGAUUAAAAUCUCAUUGCAUGCCCCACAAGGUAUGCAAUAAAA --.................(((((((....(((((((.........))))))))))))))......((((((((((......)))))))))).... ( -21.40) >DroSim_CAF1 51041 96 + 1 AGUCCAGCUAAAUAAUUAAUUAAUCCGGCUUUUGUUUCCUUACAACAGACAAGGGAUUAAAAUCUCAUUGCAUGCCCCACAAGGUAUGCAAUAAAA ...................(((((((.(((.((((......)))).)).)...)))))))......((((((((((......)))))))))).... ( -23.10) >DroEre_CAF1 46563 96 + 1 AGUCCAGCUUAAUAAUUAAAUAAUCCGGCUUUUCUUUCCUUACCAAAGACAAGGGAUUAAAAUCUCAUUGCAUGCCCCCCGAAGUAUGCAAUAAAA ....................((((((..(((.(((((.......))))).))))))))).......(((((((((........))))))))).... ( -18.70) >DroYak_CAF1 47615 96 + 1 AGUUCAGCAAAAUAAUUAAUUAAUCCGGGUUUUGUUGCCUUACCAAAGACAUGGGAUUAAAAUCUCAUUGCAUGCCCCACAAGGUAUGCAAUAAAA ...................(((((((.((((((............))))).).)))))))......((((((((((......)))))))))).... ( -23.10) >DroAna_CAF1 46148 83 + 1 -------AAAAGAAACUAAUUAAUCCGCA------ACCCCAACCAAGGACAUGGGAUUAAAAUCUCGUUUAGUUCCGCCUGAAAUAUACAAUAAAA -------....(.(((((((((((((.((------...((......))...)))))))))........)))))).).................... ( -10.70) >consensus AGUCCAGCUAAAUAAUUAAUUAAUCCGGCUUUUGUUUCCUUACCAAAGACAAGGGAUUAAAAUCUCAUUGCAUGCCCCACAAGGUAUGCAAUAAAA ...................(((((((...........................)))))))......((((((((((......)))))))))).... (-12.59 = -13.65 + 1.06)


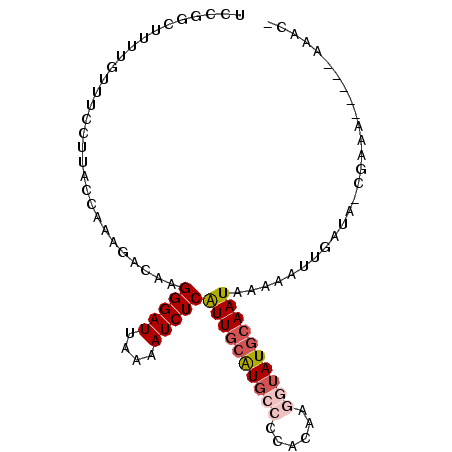
| Location | 10,811,986 – 10,812,076 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -12.19 |
| Energy contribution | -13.08 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10811986 90 + 22407834 UCCGGCUUUCGUUUCGUUACCAAAGACAAGGGAUUAAAAUCUCAUUGCAUGGCCCACAAGGUAUGCAAUAAAGAUUGAUACCAAAA-----AAAC- ...((...(((..(((((......)))..(((((....)))))((((((((.((.....))))))))))...)).)))..))....-----....- ( -17.30) >DroSec_CAF1 45628 89 + 1 UCCGGCUUUUGUUUCCUUACCACAGACAAGGGAUUAAAAUCUCAUUGCAUGCCCCACAAGGUAUGCAAUAAAAAUUGAUC-CGAAA-----AAAC- .......(((((((.........)))))))(((((((......((((((((((......)))))))))).....))))))-)....-----....- ( -22.20) >DroSim_CAF1 51064 89 + 1 UCCGGCUUUUGUUUCCUUACAACAGACAAGGGAUUAAAAUCUCAUUGCAUGCCCCACAAGGUAUGCAAUAAAAAUUGAUC-CGAAA-----AAAA- ..(((((.((((......)))).)).((((((((....)))))((((((((((......)))))))))).....)))..)-))...-----....- ( -23.20) >DroEre_CAF1 46586 89 + 1 UCCGGCUUUUCUUUCCUUACCAAAGACAAGGGAUUAAAAUCUCAUUGCAUGCCCCCCGAAGUAUGCAAUAAAAAUUGAUA-CGGAA-----AAAA- ((((.....(((((.......)))))((((((((....)))))(((((((((........))))))))).....)))...-)))).-----....- ( -20.70) >DroYak_CAF1 47638 89 + 1 UCCGGGUUUUGUUGCCUUACCAAAGACAUGGGAUUAAAAUCUCAUUGCAUGCCCCACAAGGUAUGCAAUAAAAAUUGAUA-CGAAA-----AAAC- ..(((((......)))....(((......(((((....)))))((((((((((......)))))))))).....)))...-))...-----....- ( -20.90) >DroAna_CAF1 46164 89 + 1 UCCGCA------ACCCCAACCAAGGACAUGGGAUUAAAAUCUCGUUUAGUUCCGCCUGAAAUAUACAAUAAAAACUGAAC-CUGACCACUGACAUC ......------...........((....(((((....)))))((((((((.....((.......)).....))))))))-....))......... ( -10.80) >consensus UCCGGCUUUUGUUUCCUUACCAAAGACAAGGGAUUAAAAUCUCAUUGCAUGCCCCACAAGGUAUGCAAUAAAAAUUGAUA_CGAAA_____AAAC_ .............................(((((....)))))((((((((((......))))))))))........................... (-12.19 = -13.08 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:59:11 2006