
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,762,504 – 10,762,613 |
| Length | 109 |
| Max. P | 0.685958 |

| Location | 10,762,504 – 10,762,613 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -21.28 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
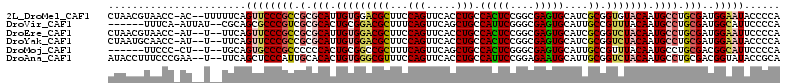
>2L_DroMel_CAF1 10762504 109 + 22407834 CUAACGUAACC-AC--UUUUUCAGUUCCCGCCGCGCAUUGUGGACGCUUCCAGUUCACCUGCCACUCCGGCGAGUGCAUCGCGGUGUACAAUGCCUGCGAUGGAAUACCCCA ...........-..--.......(((((....((((...((((((.......)))))).((((.....)))).))))(((((((.((.....))))))))))))))...... ( -32.30) >DroVir_CAF1 26096 103 + 1 ------UUUCA-AUUAU--CGCAGCGCCCGUCGCGCACUGCGGACGCUUUCAGUUCAGCUGCCAUUCGGGCGAGUGCAUUGCCGUUUACAAUGCCUGCGAUGGCAUUCCCCA ------.....-.((((--(((((((((((....(((..((((((.......)))).)))))....))))))...((((((.......)))))))))))))))......... ( -37.00) >DroEre_CAF1 17170 107 + 1 CUAACGUAACC-AU--U--UUCAGUUCCCGCCGCGCAUUGUGGACGCUUCCAGUUCACCUGCCACUCCGGCGAGUGCAUCGCGGUCUACAAUGCCUGCGAUGGAAUUCCCCA ...........-..--.--...(((((((((.(.(((((((((((((.....((.(((.((((.....)))).))).)).)).)))))))))))).)))..))))))..... ( -34.90) >DroYak_CAF1 17581 107 + 1 CUAAUGCAACC-AU--U--UUCAGUUCCCGCCGCGCAUUGUGGACGCUUCCAGUUCACCUGCCACUCCGGCGAGUGCAUCGCGGUCUACAAUGCCUGCGAUGGAAUACCCCA ...........-..--.--....((((((((.(.(((((((((((((.....((.(((.((((.....)))).))).)).)).)))))))))))).)))..)))))...... ( -34.90) >DroMoj_CAF1 26130 101 + 1 ------UUCCC-CU--U--UGCAGUGCCCGCCCCCCACUGCGGCCGCUUUCAGUUCAGCUGCCACUCGGGCGAGUGCAUUGCCGUUUACAAUGCCUGCGACGGCAUUCCCCA ------.....-..--.--.(((((((((((((......(.(((.(((........))).))).)..))))).).))))))).......((((((......))))))..... ( -35.00) >DroAna_CAF1 17320 108 + 1 AUACCUUUCCCGAA--U--UUCAGCUCCCAUUGCACACUGUGGGCGUUUCCAGUUCACCUGCCAUUCCGGAGAAUGCAUUGCGGUCUACAAUGCCUGCGACGGUAUACCGCA .....(((((.(((--(--...(((.(((((........))))).)))..(((.....)))..)))).)))))......((((((.....(((((......))))))))))) ( -27.90) >consensus CUAACGUAACC_AU__U__UUCAGUUCCCGCCGCGCACUGUGGACGCUUCCAGUUCACCUGCCACUCCGGCGAGUGCAUCGCGGUCUACAAUGCCUGCGAUGGAAUACCCCA .......................((((((((.(.(((.(((((((((...(((.....))).(((((....)))))....)).))))))).)))).)))..)))))...... (-21.28 = -21.78 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:49 2006