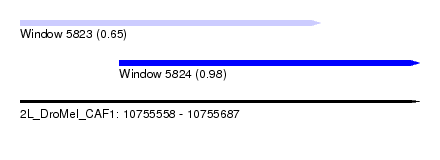
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,755,558 – 10,755,687 |
| Length | 129 |
| Max. P | 0.981572 |

| Location | 10,755,558 – 10,755,655 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.74 |
| Mean single sequence MFE | -19.61 |
| Consensus MFE | -11.84 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
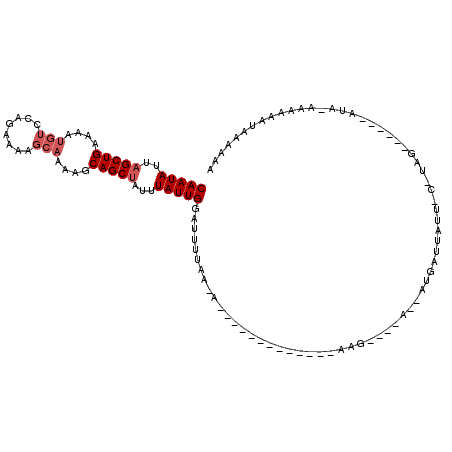
>2L_DroMel_CAF1 10755558 97 + 22407834 GCUGUAAGACAGAAUUUUCAUUUUUUGUUCAACAAUAUUAGCUGAAAAUGUCCAGAAAAGCAAAAGCAGCCAUUUAUUGGAUUUUAAG-A----------UUUAGG----AU .(((.....))).(((((((.((..((((....))))..)).)))))))((((......((....))..(((.....)))........-.----------....))----)) ( -14.00) >DroSec_CAF1 10651 90 + 1 GCUGUAAGACAGA-UUUUCAUUUUUUGUUCAACAAUAUUAGCUGAAAAUGUCCAGAAAAGCAAAAGCAGCUAUUUAUUGGAUCUAU-----------------UGG----AU .......((((((-.........))))))...(((((.((((((....(((........)))....))))))..))))).......-----------------...----.. ( -14.90) >DroSim_CAF1 9866 89 + 1 GCUGUAAGACAGAAUUUUCAUUUUUUGUUCAACAAUAUUAGCUGAAAAUGUCCAGAAAAGCAAAAGCAGCUAUUUAUUGAAUUUCA-----------------GAG------ .(((...(((((((........)))))))...(((((.((((((....(((........)))....))))))..))))).....))-----------------)..------ ( -18.00) >DroEre_CAF1 9859 104 + 1 GGUGUAAAACAGCAUUUUCAUUUUGUGUUAAACAAUAUUAGCUGAAGUUGUCCAGA------AUACCAGCUC-UUAUUGGUAUUUGAC-AAAUAUAUUAUACGAAUCCCCAA .((((((..((((.........((((.....)))).....))))...(((((..((------(((((((...-...))))))))))))-)).....)))))).......... ( -22.14) >DroYak_CAF1 10144 110 + 1 GGUGUAAAACAGCAUUUUCAUUUGUUGUUCAACAAUAUUAGCUGAAAAUGUUGAGAGA--CAGUGACAGCUUUUUGUUGGCUUUUAACCAAAAAUAUUAAUCUAAUACCAAC (((((..(((((((........)))))))((((((....(((((....((((....))--))....)))))..)))))).........................)))))... ( -29.00) >consensus GCUGUAAGACAGAAUUUUCAUUUUUUGUUCAACAAUAUUAGCUGAAAAUGUCCAGAAAAGCAAAAGCAGCUAUUUAUUGGAUUUUAA__A_____________AAG____AU .......(((((((........)))))))...(((((..(((((....(((........)))....)))))...)))))................................. (-11.84 = -12.80 + 0.96)


| Location | 10,755,590 – 10,755,687 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 54.05 |
| Mean single sequence MFE | -15.03 |
| Consensus MFE | -7.23 |
| Energy contribution | -8.57 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10755590 97 + 22407834 CAAUAUUAGCUGAAAAUGUCCAGAAAAGCAAAAGCAGCCAUUUAUUGGAUUUUAAGA----------UUUAGG----AUUAUGAUUUUCUCAUAG------GUA-AAAAAAUCAAAAA (((((...((((....(((........)))....))))....)))))........((----------(((...----.((((((.....))))))------...-...)))))..... ( -14.50) >DroSim_CAF1 9898 82 + 1 CAAUAUUAGCUGAAAAUGUCCAGAAAAGCAAAAGCAGCUAUUUAUUGAAUUUCA----------------GAG--------GUAAAAAU----AA------A-G-AAAAAACAAAAAA (((((.((((((....(((........)))....))))))..))))).......----------------...--------........----..------.-.-............. ( -12.10) >DroEre_CAF1 9891 111 + 1 CAAUAUUAGCUGAAGUUGUCCAGA------AUACCAGCUC-UUAUUGGUAUUUGACAAAUAUAUUAUACGAAUCCCCAAAAUGAUUCUUACGUAGUAAAAGAUCAACAAAAUGAAGAA ........(.(((..(((((..((------(((((((...-...))))))))))))))...((((((..(((((........)))))....)))))).....))).)........... ( -18.50) >consensus CAAUAUUAGCUGAAAAUGUCCAGAAAAGCAAAAGCAGCUAUUUAUUGGAUUUUAA_A_____________AAG____A__AUGAUUAUU_C_UAG______AUA_AAAAAAUAAAAAA (((((..(((((....(((........)))....)))))...)))))....................................................................... ( -7.23 = -8.57 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:48 2006