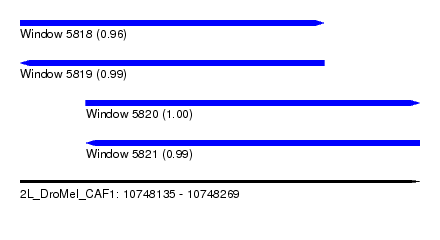
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,748,135 – 10,748,269 |
| Length | 134 |
| Max. P | 0.999843 |

| Location | 10,748,135 – 10,748,237 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -20.42 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10748135 102 + 22407834 CUUGCCACAUCU------GAUG------------UUGCAGCUUUGCAUAAAUAUUAUGUUGAAAUGCCUAUGGCAAUGCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGCCAGCAGC .((((.......------....------------..(((..((.(((((.....))))).))..)))...((((..((((....).((((((((....)))))))))))..)))))))). ( -31.30) >DroPse_CAF1 2676 114 + 1 CUUGCCACAUCUGAAGCUGACGCUGACGCUGGGGUUGGGGCUUUGCAUAAGUAUUAUGUUGAAAUGGCUCCAGCAAUGCGGCCGUAACAUGCAACAUGUUGCAUGUUCAGCGGC------ ..............(((....)))..(((....(((((((((..(((((.....)))))......)))))))))...)))((((((((((((((....)))))))))..)))))------ ( -42.00) >DroEre_CAF1 2548 96 + 1 CUUGCCACAUCU------GAUG------------UUGCAGCUUUGCAUAAACACUAUGUUGAAAUGCCUAUGGCAAUGCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGGC------ ...(((.(....------..((------------((((.((((.(((((.....))))).))..((((...))))..)).))))))((((((((....))))))))...).)))------ ( -31.20) >DroYak_CAF1 2589 96 + 1 CUUGCCACAUCU------GAUG------------UUGCAGCUUUGCAUAAAUAUUAUGUUGAAAUGCCUAUGGCAAUGCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGGC------ ...(((.(....------..((------------((((.((((.(((((.....))))).))..((((...))))..)).))))))((((((((....))))))))...).)))------ ( -31.00) >DroAna_CAF1 2758 91 + 1 CUUGCCACAUCU------AAAG------------UUGCGAUUUUGCAUAAAUAUUGUGUUGAAAUGCCAACGGCGAUGCGGUAACAACAUGCAACAUGUUGCAUGUGCA----------- .(((((.((((.------...(------------(((((.(((.(((((.....))))).))).)).))))...)))).)))))..((((((((....))))))))...----------- ( -29.20) >DroPer_CAF1 2619 108 + 1 CUUGCCACAUCU------GACGCUGACGCUGGGGCUGGGGCUUUGCAUAAGUAUUAUGUUGAAAUGGCUCCAGCAAUGCGGCCGUAACAUGCAACAUGUUGCAUGUUCAGCGGC------ ............------..(((((((((....(((((((((..(((((.....)))))......)))))))))...)))......((((((((....))))))))))))))..------ ( -43.60) >consensus CUUGCCACAUCU______GAUG____________UUGCAGCUUUGCAUAAAUAUUAUGUUGAAAUGCCUACGGCAAUGCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGGC______ .(((((.(((...............................((.(((((.....))))).))..(((.....)))))).)))))..((((((((....)))))))).............. (-20.42 = -20.45 + 0.03)

| Location | 10,748,135 – 10,748,237 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10748135 102 - 22407834 GCUGCUGGCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGCAUUGCCAUAGGCAUUUCAACAUAAUAUUUAUGCAAAGCUGCAA------------CAUC------AGAUGUGGCAAG ((.....))((..(((((((((....))))))))((((((.((.((((.(((((.(((.......))).))))))))).)).))))------------))..------....)..))... ( -34.20) >DroPse_CAF1 2676 114 - 1 ------GCCGCUGAACAUGCAACAUGUUGCAUGUUACGGCCGCAUUGCUGGAGCCAUUUCAACAUAAUACUUAUGCAAAGCCCCAACCCCAGCGUCAGCGUCAGCUUCAGAUGUGGCAAG ------((((((((((((((((....))))))))).)))).((((..(((((((........((((.....))))................((....))....))))))))))))))... ( -36.60) >DroEre_CAF1 2548 96 - 1 ------GCCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGCAUUGCCAUAGGCAUUUCAACAUAGUGUUUAUGCAAAGCUGCAA------------CAUC------AGAUGUGGCAAG ------(((((...((((((((....))))))))((((((.((.((((.(((((((((.......))))))))))))).)).))))------------))..------....)))))... ( -41.20) >DroYak_CAF1 2589 96 - 1 ------GCCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGCAUUGCCAUAGGCAUUUCAACAUAAUAUUUAUGCAAAGCUGCAA------------CAUC------AGAUGUGGCAAG ------(((((...((((((((....))))))))((((((.((.((((.(((((.(((.......))).))))))))).)).))))------------))..------....)))))... ( -34.90) >DroAna_CAF1 2758 91 - 1 -----------UGCACAUGCAACAUGUUGCAUGUUGUUACCGCAUCGCCGUUGGCAUUUCAACACAAUAUUUAUGCAAAAUCGCAA------------CUUU------AGAUGUGGCAAG -----------.((((((((((....))))))).)))..(((((((...(((((....)))))..........(((......))).------------....------.))))))).... ( -26.90) >DroPer_CAF1 2619 108 - 1 ------GCCGCUGAACAUGCAACAUGUUGCAUGUUACGGCCGCAUUGCUGGAGCCAUUUCAACAUAAUACUUAUGCAAAGCCCCAGCCCCAGCGUCAGCGUC------AGAUGUGGCAAG ------((((((((((((((((....))))))))).))))(((..((((((.((........((((.....))))..........)).))))))...)))..------......)))... ( -37.67) >consensus ______GCCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGCAUUGCCAUAGGCAUUUCAACAUAAUAUUUAUGCAAAGCCGCAA____________CAUC______AGAUGUGGCAAG ..............((((((((....))))))))....(((((((((((...)))..................(((......)))........................))))))))... (-22.48 = -22.95 + 0.47)
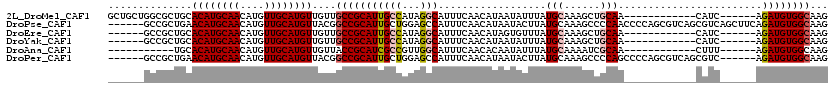

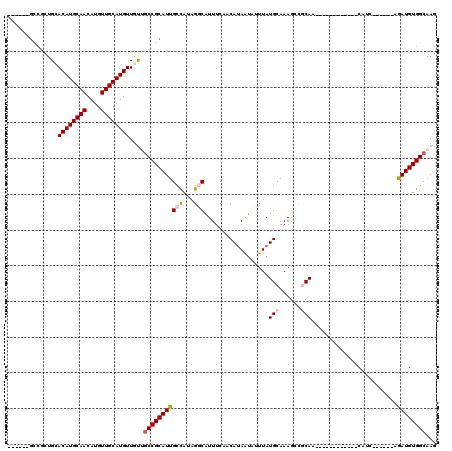
| Location | 10,748,157 – 10,748,269 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.50 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10748157 112 + 22407834 CUUUGCAUAAAUAUUAUGUUGAAAUGCCUAUGGCAAUGCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGCCAGCAGCAGCAGCUGGCCCAGCAGCAACAACAAAGUCGG- (((((...........(((((...(((...((((..((((....).((((((((....)))))))))))..))))))).)))))((((......)))).....)))))....- ( -38.80) >DroPse_CAF1 2716 98 + 1 CUUUGCAUAAGUAUUAUGUUGAAAUGGCUCCAGCAAUGCGGCCGUAACAUGCAACAUGUUGCAUGUUCAGCGGC------------CAG---AAAAACAACAACAAACUACAC ..........(((.(.(((((.....((....)).....(((((((((((((((....)))))))))..)))))------------)..---........))))).).))).. ( -31.30) >DroEre_CAF1 2570 106 + 1 CUUUGCAUAAACACUAUGUUGAAAUGCCUAUGGCAAUGCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGGC------AACAGCUGGCCCAGCAGCAGCAGCAAAAGCGG- .((((((((.....)))((((...((((...)))).((((((....((((((((....)))))))).(((((..------..).)))))))..))).)))).))))).....- ( -34.50) >DroYak_CAF1 2611 91 + 1 CUUUGCAUAAAUAUUAUGUUGAAAUGCCUAUGGCAAUGCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGGC------AACAGCUGGCCCAGCAG---------------- ................(((((...((((...))))....(((....((((((((....)))))))).(((((..------..).)))))))))))).---------------- ( -30.60) >DroPer_CAF1 2653 98 + 1 CUUUGCAUAAGUAUUAUGUUGAAAUGGCUCCAGCAAUGCGGCCGUAACAUGCAACAUGUUGCAUGUUCAGCGGC------------CAG---AAAAACAACAACAAGCUACAC ..........(((.(.(((((.....((....)).....(((((((((((((((....)))))))))..)))))------------)..---........))))).).))).. ( -31.30) >consensus CUUUGCAUAAAUAUUAUGUUGAAAUGCCUAUGGCAAUGCGGCAACAACAUGCAACAUGUUGCAUGUGCAGCGGC______A_CAGCUGGCCCAGCAGCAACAACAAACUACA_ ...(((...........((((...(((.....)))...))))....((((((((....))))))))...)))......................................... (-16.62 = -17.02 + 0.40)

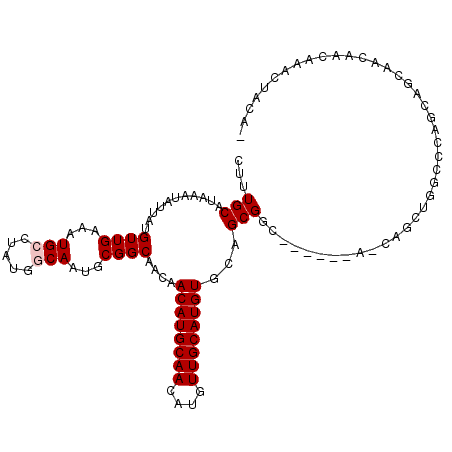
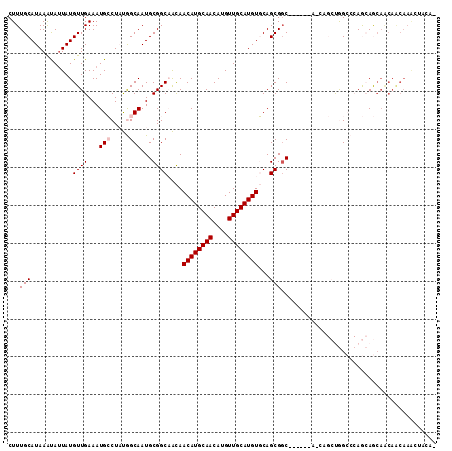
| Location | 10,748,157 – 10,748,269 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10748157 112 - 22407834 -CCGACUUUGUUGUUGCUGCUGGGCCAGCUGCUGCUGCUGGCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGCAUUGCCAUAGGCAUUUCAACAUAAUAUUUAUGCAAAG -....(((((((((((((((.(.((((((.......)))))).).)))...))))))......((((((.((((...........))))...)))))).........)))))) ( -40.00) >DroPse_CAF1 2716 98 - 1 GUGUAGUUUGUUGUUGUUUUU---CUG------------GCCGCUGAACAUGCAACAUGUUGCAUGUUACGGCCGCAUUGCUGGAGCCAUUUCAACAUAAUACUUAUGCAAAG (((((((.(((((..((..((---(((------------((((...(((((((((....))))))))).)))))((...)).))))..))..)))))....)).))))).... ( -32.30) >DroEre_CAF1 2570 106 - 1 -CCGCUUUUGCUGCUGCUGCUGGGCCAGCUGUU------GCCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGCAUUGCCAUAGGCAUUUCAACAUAGUGUUUAUGCAAAG -..((....)).((((((....)).))))...(------((.((.((((((((((....))))))).))).)).)))((((.(((((((((.......))))))))))))).. ( -35.10) >DroYak_CAF1 2611 91 - 1 ----------------CUGCUGGGCCAGCUGUU------GCCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGCAUUGCCAUAGGCAUUUCAACAUAAUAUUUAUGCAAAG ----------------.((((.(((.......(------((.((.((((((((((....))))))).))).)).)))..)))...))))......((((.....))))..... ( -26.50) >DroPer_CAF1 2653 98 - 1 GUGUAGCUUGUUGUUGUUUUU---CUG------------GCCGCUGAACAUGCAACAUGUUGCAUGUUACGGCCGCAUUGCUGGAGCCAUUUCAACAUAAUACUUAUGCAAAG ..(((...(((((..((..((---(((------------((((...(((((((((....))))))))).)))))((...)).))))..))..)))))...))).......... ( -31.60) >consensus _CCGAGUUUGUUGUUGCUGCUGGGCCAGCUG_U______GCCGCUGCACAUGCAACAUGUUGCAUGUUGUUGCCGCAUUGCCAUAGGCAUUUCAACAUAAUAUUUAUGCAAAG .......................................((.((.((((((((((....))))))).))).)).)).((((.(((((.(((.......))).))))))))).. (-17.02 = -17.78 + 0.76)

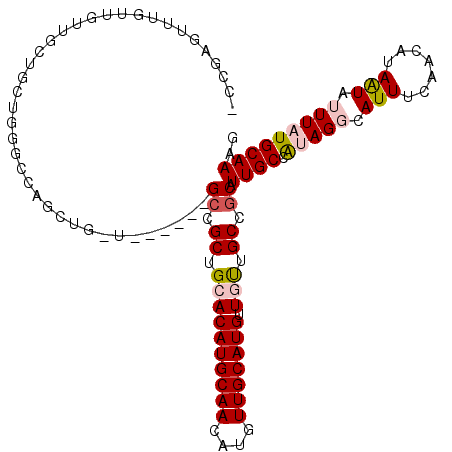

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:45 2006