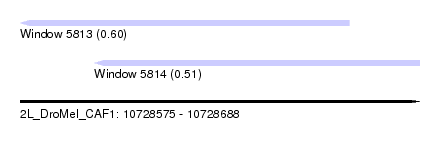
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,728,575 – 10,728,688 |
| Length | 113 |
| Max. P | 0.604962 |

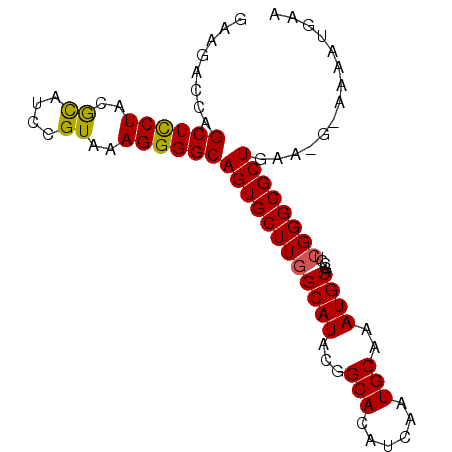
| Location | 10,728,575 – 10,728,668 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -18.03 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10728575 93 - 22407834 CCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUAAAUCAAAUAUGAAAUUG------GUUAUAAAUUAUAUAUA----- .(((...(((((.(((((........))))).....(((....))))))))..)))...((((((........)))------)))..............----- ( -22.90) >DroPse_CAF1 2314 95 - 1 CCGUGAAGGGGCAGUGCUUGGCAUAAGGCACAUCUAUGCAAAUGCAGCCACGGGCGCUGGAAG---CAUGUGUUUUUGAUACAUUUGAAAUUAAAAA-A----- ((((.....(((.((((((......)))))).....(((....))))))))))((((((....---)).))))(((((((.........))))))).-.----- ( -24.10) >DroSim_CAF1 1014 89 - 1 CCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUGA----AAAAUGAAUUUG------GUUAUAAAUUAUAUAUU----- .(((...(((((.(((((........))))).....(((....))))))))..)))....----......((((((------....)))))).......----- ( -21.60) >DroEre_CAF1 2455 98 - 1 CCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAACCUCGGGCGCUAAAAGAAAACGGAAAUUG------GUUAAAAAUUAUAUACAAAUGA ((((........((((((((((((...(((......)))..)))).....))))))))........))))......------...................... ( -18.89) >DroYak_CAF1 1748 98 - 1 CCGUAAAGGGGCAGUGCUUCGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUGAUCGGAAACGGAAAUAG------GUUAGAAACUGAUUGAAUGUUG .(((...(((((.(((((........))))).....(((....))))))))..)))....(((....))).....(------(((((...))))))........ ( -26.30) >DroPer_CAF1 2316 95 - 1 CCGUGAAGGGGCAGUGCUUGGCAUAAGGCACAUCUAUGCAAAUGCAGCCACGGGCGCUGGAAG---CAUGUGUUUUUGAUACAUUUGAAAUUAAAGA-A----- ((((.....(((.((((((......)))))).....(((....))))))))))((((((....---)).))))(((..(.....)..))).......-.----- ( -23.90) >consensus CCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUGAAAG_AAAAUGAAAUUG______GUUAGAAAUUAAAUA_A_____ .(((...(((((.(((((........))))).....(((....))))))))..)))................................................ (-18.03 = -18.53 + 0.50)

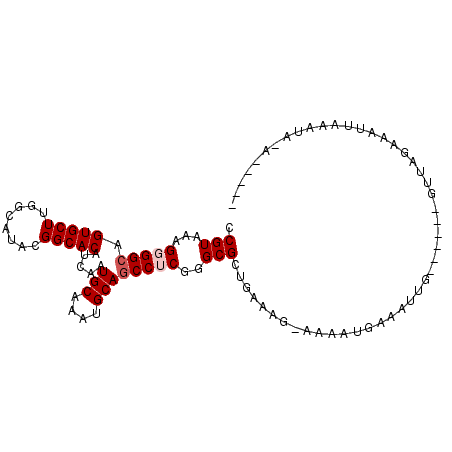
| Location | 10,728,596 – 10,728,688 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.83 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10728596 92 - 22407834 GAAGACCAGCUCCUACGCAUCCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUAAAUCAAAUAUGAA ...((..((((((((((....))))..(((((.(((((........))))).....(((....))))))))))).)))...))......... ( -25.50) >DroPse_CAF1 2340 89 - 1 GGUGGCCAGCUUCUACAUAUCCGUGAAGGGGCAGUGCUUGGCAUAAGGCACAUCUAUGCAAAUGCAGCCACGGGCGCUGGAAG---CAUGUG .((..(((((.(((......((.....))(((.((((((......)))))).....(((....))))))..))).)))))..)---)..... ( -31.40) >DroSec_CAF1 2314 88 - 1 GAAGACCAGCUCCUACGCAUCCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUGA----AAAAUGAA ......(((((((((((....))))..(((((.(((((........))))).....(((....))))))))))).)))).----........ ( -28.40) >DroSim_CAF1 1035 88 - 1 GAAGACCAGCUCCUACGCAUCCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUGA----AAAAUGAA ......(((((((((((....))))..(((((.(((((........))))).....(((....))))))))))).)))).----........ ( -28.40) >DroEre_CAF1 2481 92 - 1 GAAGACCAGCUCCUACGCGUCCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAACCUCGGGCGCUAAAAGAAAACGGAA .....((.((((((..((....))..))))))((((((((((((...(((......)))..)))).....))))))))..........)).. ( -27.50) >DroYak_CAF1 1774 92 - 1 GAAGACCAGCUCCUACGCGUCCGUAAAGGGGCAGUGCUUCGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUGAUCGGAAACGGAA ......(((((((((((....))))..(((((.(((((........))))).....(((....))))))))))).)))).(((....))).. ( -32.70) >consensus GAAGACCAGCUCCUACGCAUCCGUAAAGGGGCAGUGCUUGGCAUACGGCACAUCAAUGCAAAUGCAGCCUCGGGCGCUGAA_G_AAAAUGAA ........((((((..((....))..))))))((((((((((((...(((......)))..)))).....)))))))).............. (-24.08 = -23.83 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:39 2006