| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,709,447 – 10,709,592 |
| Length | 145 |
| Max. P | 0.819870 |

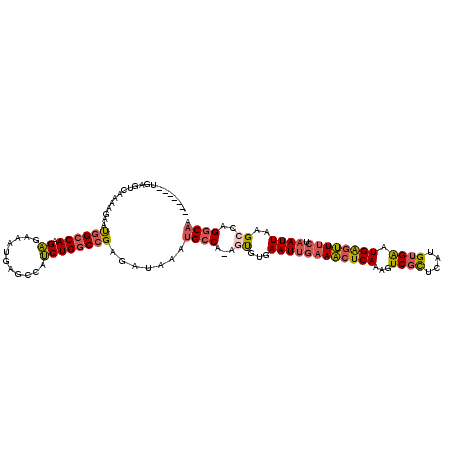
| Location | 10,709,447 – 10,709,565 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
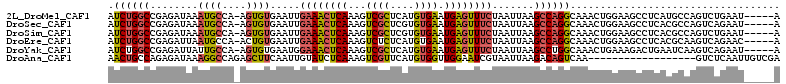
>2L_DroMel_CAF1 10709447 118 + 22407834 UGCUGAAUGAGUCAAAAGAAUGGCCAAGAGAAAUGAGCCAUCUGGCCGAGAUAAAUGCCA-AGUGUGAAUUGAAACUCAAAGUCGCUCAUGUGAAUGAGUUUCUAAUUAAGCCAGGCAA ....................((((((.((...........)))))))).......((((.-.((...((((((((((((...((((....)))).)))))))).))))..))..)))). ( -32.40) >DroSec_CAF1 40385 104 + 1 --------------AAAGAAUGGCCAAGAGAAAUGAGCCAUCUGGCCGAGAUAAAUGCCA-AGUGUGAAUUGAAACUCAAAGUCGCUCGUGUGAAUGAGUUUCUAAUUAAGCCAGGCAA --------------......((((((.((...........)))))))).......((((.-.((...((((((((((((...((((....)))).)))))))).))))..))..)))). ( -32.40) >DroSim_CAF1 41360 104 + 1 --------------AAAGAAUGGCCAAGAGAAAUGAGCCAUCUGGCCGAGAUAAAUGCCA-AGUGUGAAUUGAAACUCAAAGUCGCUCGUGUGAAUGAGUUUCUAAUUAAGCCAGGCAA --------------......((((((.((...........)))))))).......((((.-.((...((((((((((((...((((....)))).)))))))).))))..))..)))). ( -32.40) >DroEre_CAF1 41595 115 + 1 ---UAAAUGAGUCAAAAGAAUGGCCAAGAGAAAUGAGCCAUCUGGCCGAGAUUAAUGCCA-ACUGUGAAUUGAAACUCAAAGUCUCUCAUGUGAAUGAGUUUCUAAUUAAGCCAGGCAA ---.................((((((.((...........)))))))).......((((.-.((...((((((((((((...((.(....).)).)))))))).)))).))...)))). ( -26.90) >DroYak_CAF1 39745 115 + 1 ---UAAAUGACUCAAAAGAAUGGCCAAGAGAAAUGAGCCAUCUGGCCGAGAUUAUUGCCA-AGUGUGAAUGGAAACUCAAAGUCGCUCAUGUGAAUGAGUUUCUAAUUAAGCCUGGCAA ---..((((((((...(((.((((((.......)).)))))))....))).)))))((((-.((.(((.((((((((((...((((....)))).)))))))))).))).)).)))).. ( -38.60) >DroAna_CAF1 40594 112 + 1 -------UGAGACAAAAAAGCGGCCAAGUAAAAUAAGUCAACUGCCAGAGAUAAAGGCCAGAGCUUCAAUUGUAUCUCAAAGUCGUUCAUGUGGUUGGAAUCGUAAUUAAGACAGUCAA -------...(((........((((..........((....))............)))).((..((((((..(((..(......)...)))..)))))).))............))).. ( -18.05) >consensus _______UGAGUCAAAAGAAUGGCCAAGAGAAAUGAGCCAUCUGGCCGAGAUAAAUGCCA_AGUGUGAAUUGAAACUCAAAGUCGCUCAUGUGAAUGAGUUUCUAAUUAAGCCAGGCAA ....................((((((.((...........)))))))).......((((...((...((((((((((((...((((....)))).)))))))).))))..))..)))). (-21.69 = -22.28 + 0.59)

| Location | 10,709,447 – 10,709,565 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10709447 118 - 22407834 UUGCCUGGCUUAAUUAGAAACUCAUUCACAUGAGCGACUUUGAGUUUCAAUUCACACU-UGGCAUUUAUCUCGGCCAGAUGGCUCAUUUCUCUUGGCCAUUCUUUUGACUCAUUCAGCA .(((((((((......((((((((.((.(....).))...))))))))...(((....-)))..........))))))(((((.((.......)))))))................))) ( -26.60) >DroSec_CAF1 40385 104 - 1 UUGCCUGGCUUAAUUAGAAACUCAUUCACACGAGCGACUUUGAGUUUCAAUUCACACU-UGGCAUUUAUCUCGGCCAGAUGGCUCAUUUCUCUUGGCCAUUCUUU-------------- .((((.((...((((.((((((((.((.(....).))...))))))))))))....))-.))))........((((((..((......))..)))))).......-------------- ( -26.20) >DroSim_CAF1 41360 104 - 1 UUGCCUGGCUUAAUUAGAAACUCAUUCACACGAGCGACUUUGAGUUUCAAUUCACACU-UGGCAUUUAUCUCGGCCAGAUGGCUCAUUUCUCUUGGCCAUUCUUU-------------- .((((.((...((((.((((((((.((.(....).))...))))))))))))....))-.))))........((((((..((......))..)))))).......-------------- ( -26.20) >DroEre_CAF1 41595 115 - 1 UUGCCUGGCUUAAUUAGAAACUCAUUCACAUGAGAGACUUUGAGUUUCAAUUCACAGU-UGGCAUUAAUCUCGGCCAGAUGGCUCAUUUCUCUUGGCCAUUCUUUUGACUCAUUUA--- .((((.((((.((((.((((((((.((........))...))))))))))))...)))-)))))........((((((..((......))..))))))..................--- ( -26.80) >DroYak_CAF1 39745 115 - 1 UUGCCAGGCUUAAUUAGAAACUCAUUCACAUGAGCGACUUUGAGUUUCCAUUCACACU-UGGCAAUAAUCUCGGCCAGAUGGCUCAUUUCUCUUGGCCAUUCUUUUGAGUCAUUUA--- ((((((((........((((((((.((.(....).))...))))))))........))-))))))....(((((...((((((.((.......))))))))...))))).......--- ( -31.59) >DroAna_CAF1 40594 112 - 1 UUGACUGUCUUAAUUACGAUUCCAACCACAUGAACGACUUUGAGAUACAAUUGAAGCUCUGGCCUUUAUCUCUGGCAGUUGACUUAUUUUACUUGGCCGCUUUUUUGUCUCA------- ..(((((((((((...((.(((.........)))))...)))))))).....(((((...((((.........(((....).))..........)))))))))...)))...------- ( -18.31) >consensus UUGCCUGGCUUAAUUAGAAACUCAUUCACAUGAGCGACUUUGAGUUUCAAUUCACACU_UGGCAUUUAUCUCGGCCAGAUGGCUCAUUUCUCUUGGCCAUUCUUUUGACUCA_______ ....((((((......((((((((.((........))...))))))))............((........))))))))(((((.((.......)))))))................... (-17.53 = -17.90 + 0.37)
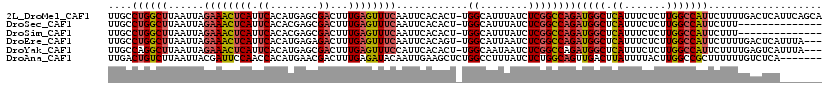
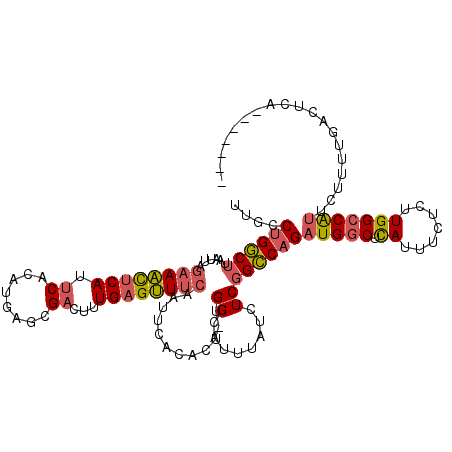
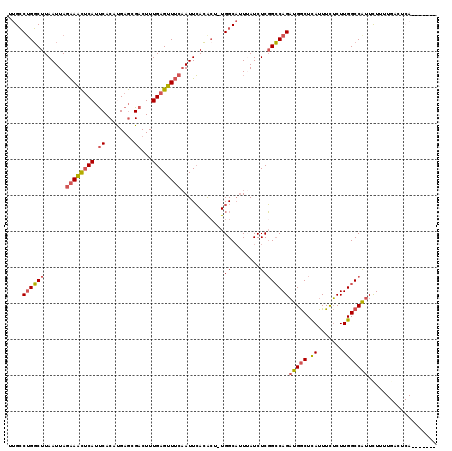
| Location | 10,709,486 – 10,709,592 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -16.78 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10709486 106 + 22407834 AUCUGGCCGAGAUAAAUGCCA-AGUGUGAAUUGAAACUCAAAGUCGCUCAUGUGAAUGAGUUUCUAAUUAAGCCAGGCAAACUGGAAGCCUCAUGCCAGUCUGAAU-----A ..(((((.(((.....((((.-.((...((((((((((((...((((....)))).)))))))).))))..))..))))....(....))))..))))).......-----. ( -31.90) >DroSec_CAF1 40410 106 + 1 AUCUGGCCGAGAUAAAUGCCA-AGUGUGAAUUGAAACUCAAAGUCGCUCGUGUGAAUGAGUUUCUAAUUAAGCCAGGCAAACUGGAAGCCUCACGCCAGUCAGAAU-----A .((((((.(.(........).-.((((((...((((((((...((((....)))).))))))))........((((.....)))).....))))))).))))))..-----. ( -32.30) >DroSim_CAF1 41385 106 + 1 AUCUGGCCGAGAUAAAUGCCA-AGUGUGAAUUGAAACUCAAAGUCGCUCGUGUGAAUGAGUUUCUAAUUAAGCCAGGCAAACUGGAAGCCUCACGCCAGUCUGAAU-----A ..(((((.(((.....((((.-.((...((((((((((((...((((....)))).)))))))).))))..))..))))....(....))))..))))).......-----. ( -31.90) >DroEre_CAF1 41631 106 + 1 AUCUGGCCGAGAUUAAUGCCA-ACUGUGAAUUGAAACUCAAAGUCUCUCAUGUGAAUGAGUUUCUAAUUAAGCCAGGCAAACUGGAAGCCUCACGCAAGUCAGAAC-----A .((((...(((..........-.((...((((((((((((...((.(....).)).)))))))).)))).))((((.....))))....)))((....))))))..-----. ( -26.00) >DroYak_CAF1 39781 106 + 1 AUCUGGCCGAGAUUAUUGCCA-AGUGUGAAUGGAAACUCAAAGUCGCUCAUGUGAAUGAGUUUCUAAUUAAGCCUGGCAAACUGAAAGACUGAAUCAAGUCAGAAU-----A .((((((...((((.((((((-.((.(((.((((((((((...((((....)))).)))))))))).))).)).)))))).(.....)....))))..))))))..-----. ( -35.50) >DroAna_CAF1 40626 94 + 1 AACUGCCAGAGAUAAAGGCCAGAGCUUCAAUUGUAUCUCAAAGUCGUUCAUGUGGUUGGAAUCGUAAUUAAGACAGUCAA------------------GUCUCAAUUGUCGA ..(.((((((((((.((((....))))......))))))...(.....)...)))).)...((((((((.((((......------------------)))).))))).))) ( -18.30) >consensus AUCUGGCCGAGAUAAAUGCCA_AGUGUGAAUUGAAACUCAAAGUCGCUCAUGUGAAUGAGUUUCUAAUUAAGCCAGGCAAACUGGAAGCCUCACGCCAGUCAGAAU_____A .((((((........((((....)))).....((((((((...((((....)))).)))))))).......))))))................................... (-16.78 = -16.73 + -0.05)

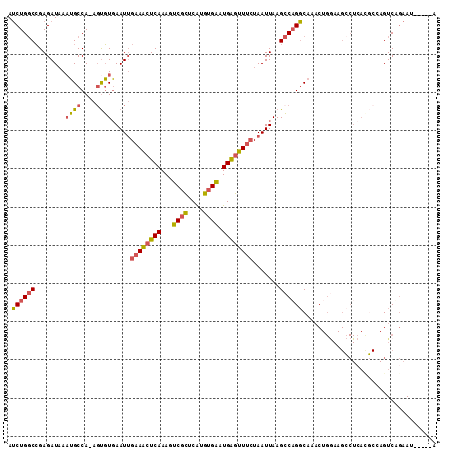
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:32 2006