
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,670,882 – 10,671,030 |
| Length | 148 |
| Max. P | 0.933291 |

| Location | 10,670,882 – 10,670,994 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -23.07 |
| Energy contribution | -23.12 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10670882 112 + 22407834 GUGCGCCGGGUGGAGAAUCAUCGGGCCAAGCUCACGGCCAAUUUGGCCAAUUACCUGGCCAUGCAGCUGCAGCGCCAG---CAGCUCCGCCAACAGAUGCAGCAGCAAAUGAUGG-- (((.(((((((((.........((((...))))..((((.....))))..))))))))))))(.((((((.......)---))))).).(((.((..(((....)))..)).)))-- ( -43.70) >DroVir_CAF1 3338 114 + 1 AUACGACGCGUAGAGAAUCACAGGGCCAAGCUGACGGCCAAUUUGGCCAGCUUCCUGGCCAUGCAGCUGCAGCGUCAG---CAGCUCCAGAAUCAAAUACAAAUGCAGACAACGCUU .......((((...((.((...((((...(((((((.......(((((((....)))))))(((....))).))))))---).))))..)).)).........((....)))))).. ( -36.10) >DroPse_CAF1 2083 112 + 1 AUCCGGCGGGUCGAGAACCACAAGGCAAAGCUGACGGCCAAUCUGGCCAGCUUCUUGGCCAUGCAGCUGCAGCGGCAG---CAGCUGCAGAACCAAAUCCAGUUGCAGAUGUCGC-- ...(((((((((((((.((....))...((((...((((.....)))))))))))))))).((((((((..(((((..---..))))).((......))))))))))..))))).-- ( -44.70) >DroGri_CAF1 2158 114 + 1 AUACGACGCGUUGAGAACCACAGGGCCAAGUUGACAGCCAAUUUGGCCAGUUUCCUGGCCAUGCAGCUGCAGCGCCAG---CAGUUCCAGAAUCAAAUACAAAUGCAGACAACGCUC .......((((((..........(((...((((.((((.....(((((((....)))))))....))))))))))).(---((....................)))...)))))).. ( -37.15) >DroMoj_CAF1 2210 117 + 1 AUACGACGCGUGGAGAAUCAACGGGCCAAGCUGACAGCCAAUUUGGCCAGCUUCCUGGCCAUGCAGCUGCAGCGUCAGCAGCAGCUCCAGAAUCAAAUUCAAAUGCAGACUCAACUA .........((.(((........((((((((((...(((.....))))))))...))))).(((((((((.((....)).)))))....((((...))))...))))..))).)).. ( -38.70) >DroPer_CAF1 2082 112 + 1 AUCCGGCGGGUCGAGAACCACAAGGCAAAGCUGACGGCCAAUCUGGCCAGCUUCUUGGCCAUGCAGCUGCAGCGGCAG---CAGCUGCAGAACCAAAUCCAAUUGCAGAUGUCGC-- ...(((((((((((((.((....))...((((...((((.....)))))))))))))))).(((((((((.......)---))))))))....................))))).-- ( -43.60) >consensus AUACGACGCGUCGAGAACCACAGGGCCAAGCUGACGGCCAAUUUGGCCAGCUUCCUGGCCAUGCAGCUGCAGCGCCAG___CAGCUCCAGAAUCAAAUACAAAUGCAGACGACGC__ ....((......(((........(((...((((.((((.....(((((((....)))))))....)))))))))))........))).....))....................... (-23.07 = -23.12 + 0.06)



| Location | 10,670,882 – 10,670,994 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -45.87 |
| Consensus MFE | -25.33 |
| Energy contribution | -24.87 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10670882 112 - 22407834 --CCAUCAUUUGCUGCUGCAUCUGUUGGCGGAGCUG---CUGGCGCUGCAGCUGCAUGGCCAGGUAAUUGGCCAAAUUGGCCGUGAGCUUGGCCCGAUGAUUCUCCACCCGGCGCAC --.((((...(((.((((((..(((..(((....))---)..))).)))))).))).((((((((...(((((.....)))))...)))))))).)))).................. ( -50.20) >DroVir_CAF1 3338 114 - 1 AAGCGUUGUCUGCAUUUGUAUUUGAUUCUGGAGCUG---CUGACGCUGCAGCUGCAUGGCCAGGAAGCUGGCCAAAUUGGCCGUCAGCUUGGCCCUGUGAUUCUCUACGCGUCGUAU ..((((.(((.(((..............((.(((((---(.......)))))).)).(((((((..(.(((((.....))))).)..))))))).)))))).....))))....... ( -41.70) >DroPse_CAF1 2083 112 - 1 --GCGACAUCUGCAACUGGAUUUGGUUCUGCAGCUG---CUGCCGCUGCAGCUGCAUGGCCAAGAAGCUGGCCAGAUUGGCCGUCAGCUUUGCCUUGUGGUUCUCGACCCGCCGGAU --.(((...(..(((..((..(((((..((((((((---(.......)))))))))..)))))((((((((((.....))))...)))))).)))))..)...))).((....)).. ( -46.60) >DroGri_CAF1 2158 114 - 1 GAGCGUUGUCUGCAUUUGUAUUUGAUUCUGGAACUG---CUGGCGCUGCAGCUGCAUGGCCAGGAAACUGGCCAAAUUGGCUGUCAACUUGGCCCUGUGGUUCUCAACGCGUCGUAU ((((((((..(((....))).........(((((..---(.((.((((((((((..(((((((....)))))))...)))))))......))))).)..))))))))))).)).... ( -48.00) >DroMoj_CAF1 2210 117 - 1 UAGUUGAGUCUGCAUUUGAAUUUGAUUCUGGAGCUGCUGCUGACGCUGCAGCUGCAUGGCCAGGAAGCUGGCCAAAUUGGCUGUCAGCUUGGCCCGUUGAUUCUCCACGCGUCGUAU ..(..(((((.((....((((...)))).((.(((...(((((((((((....)).(((((((....)))))))....))).))))))..))))))).)))))..)........... ( -42.10) >DroPer_CAF1 2082 112 - 1 --GCGACAUCUGCAAUUGGAUUUGGUUCUGCAGCUG---CUGCCGCUGCAGCUGCAUGGCCAAGAAGCUGGCCAGAUUGGCCGUCAGCUUUGCCUUGUGGUUCUCGACCCGCCGGAU --.(((...(..(((..((..(((((..((((((((---(.......)))))))))..)))))((((((((((.....))))...)))))).)))))..)...))).((....)).. ( -46.60) >consensus __GCGUCAUCUGCAUUUGGAUUUGAUUCUGGAGCUG___CUGACGCUGCAGCUGCAUGGCCAGGAAGCUGGCCAAAUUGGCCGUCAGCUUGGCCCUGUGAUUCUCCACCCGUCGUAU ..(((.....))).........((((...(.(((((...(....)...))))).)..(((((((....(((((.....)))))....)))))))................))))... (-25.33 = -24.87 + -0.47)


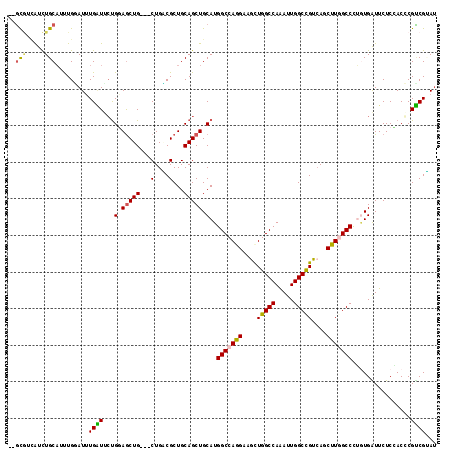
| Location | 10,670,922 – 10,671,030 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.48 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -19.82 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10670922 108 + 22407834 AUUUGGCCAAUUACCUGGCCAUGCAGCUGCAGCGCCAGCAGCUCCGCCAACAGAUGCAGCAGCAAAUGAUGG------AGGGUCUGCCGCCCAAUGUGCACCUGCCCCACGGAC ...((((((......)))))).((((.(((((((...((((..((.(((.((..(((....)))..)).)))------.))..)))))))......)))).))))......... ( -40.00) >DroVir_CAF1 3378 114 + 1 AUUUGGCCAGCUUCCUGGCCAUGCAGCUGCAGCGUCAGCAGCUCCAGAAUCAAAUACAAAUGCAGACAACGCUUCAAUCAGCUUUGCCGCCUAAUGUCCACUUGCCCCAUAGAU ...(((((((....)))))))((.((((((.......)))))).))...............(((((((..((..(((......)))..))....))))....)))......... ( -28.10) >DroPse_CAF1 2123 105 + 1 AUCUGGCCAGCUUCUUGGCCAUGCAGCUGCAGCGGCAGCAGCUGCAGAACCAAAUCCAGUUGCAGAUGUCGC------AG---CUUCCACCCAACGUCCACCUCCCCCACGGCC ....((((......((((...(((((((((.......)))))))))...))))....((((((.......))------))---)).........................)))) ( -35.00) >DroGri_CAF1 2198 114 + 1 AUUUGGCCAGUUUCCUGGCCAUGCAGCUGCAGCGCCAGCAGUUCCAGAAUCAAAUACAAAUGCAGACAACGCUCCAAUCAGCUCUGCCGCCCAAUGUUCACUUGCCUCAUAUGC ...(((((((....)))))))((.((((((.......)))))).))((((...........(((((....(((......))))))))........))))............... ( -27.21) >DroSec_CAF1 2250 108 + 1 AUUUGGCCAAUUACCUGGCCAUGCAGCUGCAGCGCCAGCAGCUCCGCCAGCAGAUGCAGCAGCAGAUGUUGG------AGGGUCUGCCGCCCAAUGUGCACCUGCCCCACGGAC ...((((((......)))))).((((.(((((((...((((..((.((((((..(((....)))..))))))------.))..)))))))......)))).))))......... ( -45.40) >DroPer_CAF1 2122 105 + 1 AUCUGGCCAGCUUCUUGGCCAUGCAGCUGCAGCGGCAGCAGCUGCAGAACCAAAUCCAAUUGCAGAUGUCGC------AG---CUUCCACCCAACGUCCACCUCCCCCACGGCC ....((((......((((...(((((((((.......)))))))))...))))......((((.......))------))---...........................)))) ( -30.80) >consensus AUUUGGCCAGCUUCCUGGCCAUGCAGCUGCAGCGCCAGCAGCUCCAGAACCAAAUACAAAUGCAGAUGACGC______AGG_UCUGCCGCCCAAUGUCCACCUGCCCCACGGAC ...(((((((....)))))))((.((((((.......)))))).)).................................................................... (-19.82 = -19.68 + -0.14)

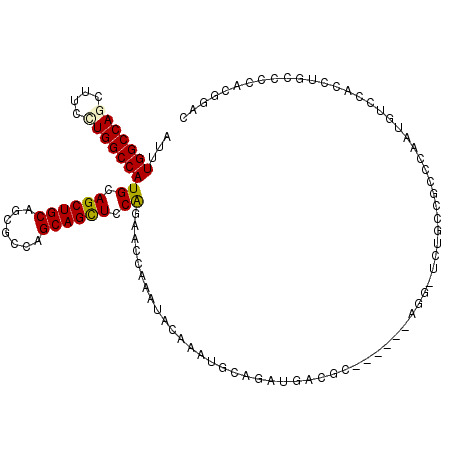
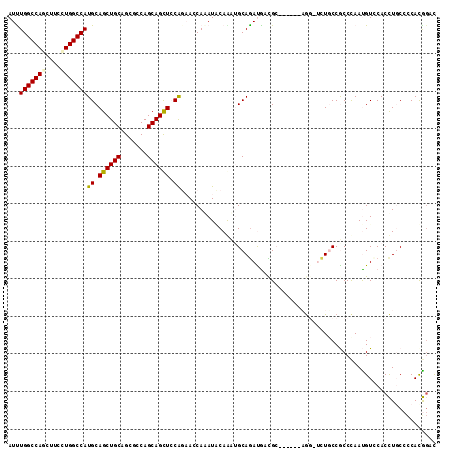
| Location | 10,670,922 – 10,671,030 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.48 |
| Mean single sequence MFE | -45.97 |
| Consensus MFE | -24.51 |
| Energy contribution | -24.43 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10670922 108 - 22407834 GUCCGUGGGGCAGGUGCACAUUGGGCGGCAGACCCU------CCAUCAUUUGCUGCUGCAUCUGUUGGCGGAGCUGCUGGCGCUGCAGCUGCAUGGCCAGGUAAUUGGCCAAAU .........((((.((((.....(.((((((..((.------(((.((..(((....)))..)).))).))..)))))).)..)))).)))).(((((((....)))))))... ( -47.90) >DroVir_CAF1 3378 114 - 1 AUCUAUGGGGCAAGUGGACAUUAGGCGGCAAAGCUGAUUGAAGCGUUGUCUGCAUUUGUAUUUGAUUCUGGAGCUGCUGACGCUGCAGCUGCAUGGCCAGGAAGCUGGCCAAAU ......(((((((((..(((....(((((((.(((......))).))).))))...)))))))).))))(.((((((.......)))))).).(((((((....)))))))... ( -43.10) >DroPse_CAF1 2123 105 - 1 GGCCGUGGGGGAGGUGGACGUUGGGUGGAAG---CU------GCGACAUCUGCAACUGGAUUUGGUUCUGCAGCUGCUGCCGCUGCAGCUGCAUGGCCAAGAAGCUGGCCAGAU ((((..(..(.(((((..(((..(((....)---))------))).))))).)..)....((((((..(((((((((.......)))))))))..)))))).....)))).... ( -45.70) >DroGri_CAF1 2198 114 - 1 GCAUAUGAGGCAAGUGAACAUUGGGCGGCAGAGCUGAUUGGAGCGUUGUCUGCAUUUGUAUUUGAUUCUGGAACUGCUGGCGCUGCAGCUGCAUGGCCAGGAAACUGGCCAAAU (((..((.(((..(....)....(.((((((..(((((..(((((.((....))..))).))..)))..))..)))))).)))).))..))).(((((((....)))))))... ( -41.70) >DroSec_CAF1 2250 108 - 1 GUCCGUGGGGCAGGUGCACAUUGGGCGGCAGACCCU------CCAACAUCUGCUGCUGCAUCUGCUGGCGGAGCUGCUGGCGCUGCAGCUGCAUGGCCAGGUAAUUGGCCAAAU .(((((.((.((((((((......((((((((....------......)))))))))))))))))).)))))(((((.......)))))....(((((((....)))))))... ( -52.70) >DroPer_CAF1 2122 105 - 1 GGCCGUGGGGGAGGUGGACGUUGGGUGGAAG---CU------GCGACAUCUGCAAUUGGAUUUGGUUCUGCAGCUGCUGCCGCUGCAGCUGCAUGGCCAAGAAGCUGGCCAGAU ((((.........(..((.(((((((....)---))------.)))).))..).......((((((..(((((((((.......)))))))))..)))))).....)))).... ( -44.70) >consensus GUCCGUGGGGCAGGUGGACAUUGGGCGGCAGA_CCU______GCGACAUCUGCAACUGCAUUUGAUUCUGGAGCUGCUGGCGCUGCAGCUGCAUGGCCAGGAAACUGGCCAAAU ..........((((((..((....((((.....................))))...)))))))).....(.((((((.......)))))).).(((((((....)))))))... (-24.51 = -24.43 + -0.08)

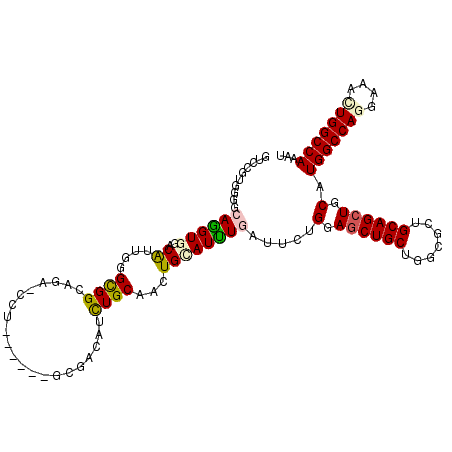
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:19 2006