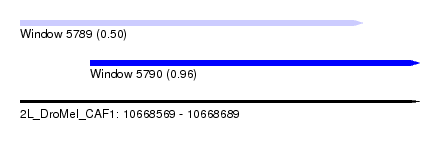
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,668,569 – 10,668,689 |
| Length | 120 |
| Max. P | 0.959535 |

| Location | 10,668,569 – 10,668,672 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -17.92 |
| Consensus MFE | -13.46 |
| Energy contribution | -14.50 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10668569 103 + 22407834 AUGGUUUUUG--CAAUUUUCAGUACUCCUUUUGUAUCAUAUGCAGAGGUAUUUAAUAAA-AUUUCCAAAAUCUAUAUUGAGUACCUGCCUGUGAUGUUAAUUAAUC ..((....((--(........)))..)).....(((((((.(((..(((((((((((..-..............)))))))))))))).))))))).......... ( -18.99) >DroSec_CAF1 21637 88 + 1 -------UU----------CAGUACUCCUUUUGUAUCAUAUGCAGAGGUAUUUAAUAAA-AUUUCCAAAAUUUACAUUCAGUACCUGCCUGUGAUGUUAAUUAAUC -------..----------..............(((((((.(((..((((((.(((.((-(((.....)))))..))).))))))))).))))))).......... ( -14.30) >DroSim_CAF1 22797 103 + 1 AUGGUUUUUCUUCAAAC--UAGUACUCCUUUUGUAUCAUAUGCAGAGGUAUUUACUUUA-AUUUCCAAAAUCAAUAUUGAGUAUCUGCCUGUGAUGUUAAUUAAUA .((((((......))))--))............(((((((.(((((((.(((......)-))..))....(((....)))...))))).))))))).......... ( -17.00) >DroEre_CAF1 24143 102 + 1 AUGGUCUUUG--CAAUUUUCAGUACUCCUUUUGUAUCAUAUGCAGAGGUAUUUCAUAAA-AUUUCCA-AAUGUAAAUUGAGUACCUGCAUGUGAUGUUAAUUAAUC ..((....((--(........)))..)).....(((((((((((..(((((((.....(-((((.(.-...).))))))))))))))))))))))).......... ( -19.60) >DroYak_CAF1 22849 93 + 1 CUGGCUUUUG--CAAUUUUCAGU-----------AACAUAUGCAGAGGUAUUUAACAAAAAUUUCCAAAAUGUAUAUUGAGUACCUGCAUGUGAUGUAAAUUAAUA ...((....)--)(((((.((..-----------..((((((((..(((((((((.....(((.....))).....))))))))))))))))).)).))))).... ( -19.70) >consensus AUGGUUUUUG__CAAUUUUCAGUACUCCUUUUGUAUCAUAUGCAGAGGUAUUUAAUAAA_AUUUCCAAAAUCUAUAUUGAGUACCUGCCUGUGAUGUUAAUUAAUC .................................(((((((.(((..((((((((((....(((.....)))....))))))))))))).))))))).......... (-13.46 = -14.50 + 1.04)


| Location | 10,668,590 – 10,668,689 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.92 |
| Mean single sequence MFE | -17.37 |
| Consensus MFE | -15.75 |
| Energy contribution | -17.50 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10668590 99 + 22407834 ACUCCUUUUGUAUCAUAUGCAGAGGUAUUUAAUAAA-AUUUCCAAAAUCUAUAUUGAGUACCUGCCUGUGAUGUUAAUUAAUCAAAUUAAAUAUGUUAUC ..........(((((((.(((..(((((((((((..-..............)))))))))))))).)))))))((((((.....)))))).......... ( -18.59) >DroSec_CAF1 21643 99 + 1 ACUCCUUUUGUAUCAUAUGCAGAGGUAUUUAAUAAA-AUUUCCAAAAUUUACAUUCAGUACCUGCCUGUGAUGUUAAUUAAUCAAAUUAAAUAUAUUAUC ..........(((((((.(((..((((((.(((.((-(((.....)))))..))).))))))))).)))))))((((((.....)))))).......... ( -14.80) >DroEre_CAF1 24164 98 + 1 ACUCCUUUUGUAUCAUAUGCAGAGGUAUUUCAUAAA-AUUUCCA-AAUGUAAAUUGAGUACCUGCAUGUGAUGUUAAUUAAUCAAAUUCAAUCUGUUACC ..........(((((((((((..(((((((.....(-((((.(.-...).)))))))))))))))))))))))........................... ( -18.70) >DroYak_CAF1 22870 89 + 1 -----------AACAUAUGCAGAGGUAUUUAACAAAAAUUUCCAAAAUGUAUAUUGAGUACCUGCAUGUGAUGUAAAUUAAUAAAUUUCAAUCUAUUACC -----------..((((((((..(((((((((.....(((.....))).....))))))))))))))))).............................. ( -17.40) >consensus ACUCCUUUUGUAUCAUAUGCAGAGGUAUUUAAUAAA_AUUUCCAAAAUGUAUAUUGAGUACCUGCAUGUGAUGUUAAUUAAUCAAAUUAAAUAUAUUACC ..........(((((((((((..((((((((((....(((.....)))....)))))))))))))))))))))........................... (-15.75 = -17.50 + 1.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:58:16 2006